Abstract
In insects, chitin is a major structural component of the cuticle and the peritrophic membrane (PM). In nature, chitin is always associated with proteins among which chitin-binding proteins (CBPs) are the most important for forming, maintaining and regulating the functions of these extracellular structures. In this study, a genome-wide search for genes encoding proteins with ChtBD2-type (peritrophin A-type) chitin-binding domains (CBDs) was conducted. A total of 53 genes encoding 56 CBPs were identified, including 15 CPAP1s (cuticular proteins analogous to peritrophins with 1 CBD), 11 CPAP3s (CPAPs with 3 CBDs) and 17 PMPs (PM proteins) with a variable number of CBDs, which are structural components of cuticle or of the PM. CBDs were also identified in enzymes of chitin metabolism including 6 chitinases and 7 chitin deacetylases encoded by 6 and 5 genes, respectively. RNA-seq analysis confirmed that PMP and CPAP genes have differential spatial expression patterns. The expression of PMP genes is midgut-specific, while CPAP genes are widely expressed in different cuticle forming tissues. Phylogenetic analysis of CBDs of proteins in insects belonging to different orders revealed that CPAP1s from different species constitute a separate family with 16 different groups, including 6 new groups identified in this study. The CPAP3s are clustered into a separate family of 7 groups present in all insect orders. Altogether, they reveal that duplication events of CBDs in CPAP1s and CPAP3s occurred prior to the evolutionary radiation of insect species. In contrast to the CPAPs, all CBDs from individual PMPs are generally clustered and distinct from other PMPs in the same species in phylogenetic analyses, indicating that the duplication of CBDs in each of these PMPs occurred after divergence of insect species. Phylogenetic analysis of these three CBP families showed that the CBDs in CPAP1s form a clearly separate family, while those found in PMPs and CPAP3s were clustered together in the phylogenetic tree. For chitinases and chitin deacetylases, most of phylogenetic analysis performed with the CBD sequences resulted in similar clustering to the one obtained by using catalytic domain sequences alone, suggesting that CBDs were incorporated into these enzymes and evolved in tandem with the catalytic domains before the diversification of different insect orders. Based on these results, the evolution of CBDs in insect CBPs is discussed to provide a new insight into the CBD sequence structure and diversity, and their evolution and expression in insects.
Keywords: Chitin-binding protein, Chitin deacetylase, Chitinase, Peritrophic matrix protein, Cuticular proteins analogous to peritrophins, Lepidoptera
1. Introduction
Chitin is a natural polysaccharide, biopolymer of N-acetyl-glucosamine, found in a large number of phyla ranging from fungi to arthropods, and is considered as one of the most abundant biomaterial on earth, next to cellulose (Cohen, 2010). In insects, chitin is an important component of extracellular structures: the cuticle, which provides mechanical support, protects insects from physical and chemical injuries, dehydration and infection by pathogens, and the peritrophic membrane (PM), which is a semi-permeable barrier lining the gut that facilitates food digestion and provides protection against physical damage, pathogen infection and dietary toxins (Kuraishi et al., 2013; Tellam et al., 1999; Terra, 2001). In nature, chitin is found invariably associated with proteins, which can be structural proteins, enzymes and antibacterial proteins (Terra and Ferreira, 2005; Willis et al., 2005). Most of these proteins are associated with chitin by non-covalent binding of one or more chitin-binding domains (CBDs) present in their protein sequences (Wang and Granados, 2001).
To date, two major types of CBPs belonging to pfam00379 and pfam01607 families have been described in insects, based on their distinct sequence characteristics of the CBDs. The insect CBPs belonging to the pfam00379 family contain the histidine-rich, cysteine-deficient chitin binding domain with the extended R&R consensus (Chitin_Bind_4), a 68 amino acid long motif exclusively found in cuticular proteins (CPR family) (Rebers and Riddiford, 1988; Willis, 2010). The number of R&R motif-containing cuticular proteins in an insect genome ranges from 32 (Apis mellifera) to more than 150 (Aedes aegypti) (Cornman and Willis, 2008; Willis, 2010) and their ability to bind chitin has been experimentally validated for some of these proteins (Rebers and Willis, 2001). The analysis of these CBPs together with other structural cuticular proteins in Manduca sexta will be presented in a separate article (Dittmer et al., in revision). The CBPs in pfam01607 family have the cysteine-containing type-2 chitin-binding domain ChtBD2, a six-cysteine motif found in insect CBPs from both the cuticle and the PM. Their capacity for binding chitin has been experimentally validated for some proteins (Arakane et al., 2003; Wang et al., 2004). The six cysteines form three intradomain disulfide bonds that are important for the stability of the proteins (Wang and Granados, 1997). However, the precise molecular mechanisms of protein-chitin interaction are still unknown (Kramer and Muthukrishnan, 2005). Initially thought to be specific to proteins from the PM (i.e. peritrophins), the ChtBD2 consensus was given the name peritrophin A-type domain (Tellam et al., 1999). Proteins with ChtBD2 domain are now known also to be in different cuticle-forming tissues since the first non-PM ChtBD2 protein was identified from the embryonic tracheae of Drosophila melanogaster (Barry et al., 1999).
Jasrapuria et al. (2010) performed an extensive analysis of the ChtBD2-containing CBPs from the genome of the Coleopteran Tribolium castaneum and classified the ChtBD2-containing CBPs into three main families. One family of ChtBD2-containing CBPs includes secreted proteins exclusively expressed in the midgut, which are likely to be associated with the PM and are named Peritrophic Matrix Proteins (PMPs). In T. castaneum, PMPs contain 1 to 14 CBDs (Jasrapuria et al., 2010). The tandem CBDs in insect CBPs has been found in Lepidoptera and Diptera earlier (Elvin et al., 1996; Shi et al., 2004; Wang and Granados, 1997; Wang et al., 2004). Such a protein structure with a large number of CBDs in tandem may facilitate chitin-protein structure formation by cross-linking chitin fibrils and meanwhile it allows the CBPs to tolerate limited proteolytic degradation without losing their function (Wang et al., 2004). In addition to PMPs, two other families of ChtBD2-containing proteins are produced in cuticle-forming tissues. These proteins are named Cuticle Proteins Analogous to Peritrophins (CPAPs). CPAPs contain either a single CBD (CPAP1 family) or three CBDs (CPAP3 family) and show no other identifiable conserved domains. CPAP3 genes in D. melanogaster have previously been named “gasp” or “obstructor” (Barry et al., 1999; Behr and Hoch, 2005).
Functional analysis of CPAP genes in T. castaneum by RNAi indicated that most CPAPs are essential and have non-redundant functions for the formation of cuticle in different parts of the insect and at different developmental stages (Jasrapuria et al., 2012). Similar observations of non-redundancy of CBPs functions in exoskeleton organization and tracheal tubulogenesis have also been described in D. melanogaster (Luschnig et al., 2006; Petkau et al., 2012). In addition to these three families of CBPs, several chitin metabolism enzymes associated with the PM and/or cuticle also contain ChtBD2 domains, such as some chitinases (CHTs) and chitin deacetylases (CDAs) (Dixit et al., 2008; Merzendorfer and Zimoch, 2003; Zhu et al., 2008). Although chitin-binding proteins (CBPs) have received increasing attention in the last two decades, molecular details of their interaction with chitin in the structural formation and functions of cuticle and PM remain to be better understood (Iconomidou et al., 2005; Jasrapuria et al., 2010).
In the present study, we performed an extensive search of the genome of the tobacco hornworm Manduca sexta (Lepidoptera: Sphingidae), which has been recently sequenced and assembled (http://agripestbase.org/manduca and http://www.ncbi.nlm.nih.gov/assembly/GCA_000262585.1), in order to identify all genes encoding ChtBD2-containing CBPs in this model Lepidopteran. We focused the analysis on the ChtBD2 sequences from CPAPs, PMPs and chitin metabolism enzymes. By performing a phylogenetic analysis of the orthologs of these genes from other insect species and based on the patterns of gene expression in different tissues and at different developmental stages, we discuss and propose a new model of evolution of CBDs in insect CBPs to provide a new insight into the CBD sequence structure and diversity, and their evolution and expression in insects.
2. Material and methods
2.1. In silico identification of genes encoding chitin-binding proteins in M. sexta genome
An extensive search of the Manduca sexta genome (Manduca base, http://agripestbase.org/manduca) was conducted to find all the proteins predicted to contain the ChtBD2 sequence consensus (pfam01607). Briefly, known chitin-binding proteins (CBPs) from other Lepidoptera were obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) and used as queries to find homologs in the M. sexta genome using BLASTp search (Altschul et al., 1990). Then, the domain structure of these CBPs was determined by searching against the Conserved Domains Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2011). The sequences of the M. sexta CBPs identified in the initial search were used as queries for a second round of BLAST search to identify additional CBPs. This step was repeated until no additional CBP genes were detected. In parallel, a BLAST search using the typical consensus domains of CBPs (ChtBD2, pfam01607) as queries was performed to search for additional genes not detected by BLAST search using entire CBP sequences.
The chitin-binding proteins identified are classified into four different classes based on their sequence similarity, domain organization and tissue/stage specificity of gene expression. They include chitin metabolism enzymes (chitinases and chitin deacetylases), cuticular proteins analogous to peritrophins (CPAPs), which are proteins expressed in cuticle-forming tissues with either one (CPAP1s) or three CBDs (CPAP3s), and peritrophic matrix proteins (PMPs), which are expressed in the midgut and can contain from one to several CBDs (Jasrapuria et al., 2010; Tellam et al., 1999).
2.2. Developmental stage- and tissue-specific gene expression
RNA-seq data were acquired from the Manduca Genome project (http://www.ncbi.nlm.nih.gov/assembly/GCA_000262585.l), which contains a total of 52 cDNA libraries prepared from eight different tissues (brain, fat bodies, midgut, Malpighian tubules, abdomen, testes, ovaries or whole larvae) from different developmental stages (from eggs to adults). Official Gene Set (OGS) 1.0 was downloaded from ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/. Manually verified sequences of chitin binding protein genes were used to replace existing ones in OGS1.0. Reads from all libraries were trimmed to 50 bp and mapped to the updated OGS1.0 using Bowtie software version 0.12.8 (Langmead et al., 2009). The FPKM (fragments per kilobase per million fragments mapped) values of each gene in each library were further calculated by RSEM software version 1.2.12 (Li and Dewey, 2011). MultiExperiment Viewer v4.9 (MeV software – TM4; available at http://www.tm4.org/mev.html) was used to generate a “heat map” representation of gene expression profiles based on FPKM values and to perform hierarchical clustering analysis (Howe et al., 2011; Saeed et al., 2003).
2.3. Phylogenetic analysis
Multiple protein sequences alignments were performed with ClustalW and then phylogenetic trees were generated by using the neighbor-joining method (Poisson substitution model; uniform substitution rate; gaps/missing data treatment: complete deletion) implemented in MEGA 6.06 software (Tamura et al., 2013). To assess the robustness of the tree branches, a bootstrap analysis of 2000 replications was carried out on the trees inferred from the neighbor joining method and bootstrap values higher than 20% are shown on each branch of all trees generated.
2.3.1. CPAP1s
Protein sequences containing one CPAP1-type CBD from M. sexta and 12 other selected species, including 15 sequences from Cole-optera, 26 from Diptera, 6 from Homoptera, 11 from Hymenoptera, 15 from Lepidoptera and 6 from Phthiraptera obtained from the NCBI database or from the Manduca base (Supplementary Table 1) were subjected to phylogenetic analysis. CPAP1s from M. sexta were named MsCPAP1-X, where X is a capital letter (from A to O) corresponding to the phylogenetic group they belong to, as described by Jasrapuria et al. (2012). For each group, a protein sequence logo was generated by using WebLogo software v3.4 (http://weblogo.threeplusone.com) (Crooks et al., 2004).
2.3.2. CPAP3s
A set of 61 proteins containing three CPAP3-type CBDs, including 8 from Coleoptera, 22 from Diptera, 5 from Homoptera, 11 from Hymenoptera, 11 from Lepidoptera and 4 from Phthiraptera (Supplementary Table 1) obtained from the NCBI database or from the Manduca base were subjected to phylogenetic analysis. CPAP3s from M. sexta were named MsCPAP3-X, where X is a capital letter (from A to E) corresponding to the phylogenetic group which they belong to, as described by Jasrapuria et al. (2012). When more than one CPAP3 belong to the same group, a number (from 1 to 4) was added to the letter.
2.3.3. PMPs
A total of 40 proteins containing PMP-type CBDs, including 11 from Coleoptera, 1 from Crustacea, 1 from Diptera, 1 from Hymenoptera and 26 from Lepidoptera found from the NCBI database or the Manduca base (Supplementary Table 1) were subjected to phylogenetic analysis. PMPs detected in the M. sexta genome were named MsPMPX, where X is a number corresponding to the number of CBDs found in the PMP (Jasrapuria et al., 2010). When two or more PMPs contain the same quantity of CBDs, a capital letter is added after the number.
2.3.4. Chitin metabolism enzymes (chitinases and chitin deacetylases)
A set of 41 CHTs containing at least one CBD, including 7 from Coleoptera, 10 from Crustacea, 12 from Diptera, 3 from Hymenoptera, 7 from Lepidoptera and 2 from Nematoda were obtained from the NCBI database or from the Manduca base (Supplementary Table 1) and subjected to phylogenetic analysis. The chitinases from M. sexta were named (MsCHT) and numbered based on the group to which they belong to (Arakane and Muthukrishnan, 2010; Tetreau et al., in review).
Similarly, 36 CDAs containing at least one CBD, including 7 from Coleoptera, 9 from Diptera, 9 from Hymenoptera and 11 from Lepidoptera were obtained from the NCBI database or from the Manduca base (Supplementary Table 1) and subjected to phylogenetic analysis. The chitin deacetylases from M. sexta were named (MsCDA) and numbered (from 1 to 5) based on the group to which they belong to (Dixit et al., 2008; Tetreau et al., in review).
3. Results & discussion
3.1. Identification of ChtBD2-containing proteins and characterization
A total of 56 chitin-binding proteins (CBPs) (encoded by 53 different genes) containing at least one ChtBD2 domain were found in the M. sexta genome, which include 15 CPAP1s, 11 CPAP3s, 17 PMPs, 6 chitinases and 7 chitin deacetylases (Table 1). The CPAP3s from M. sexta have only limited variation in size (163–296 amino acid residues in length and 18.2–33.0 kDa in molecular weight) and predicted isoelectric point (pI = 4.7–5.5). MsCPAP3-E3 is the only exception with a significantly longer size than the other CPAP3s (640 amino acid residues in length). In contrast, CPAP1s and PMPs have a much larger variation in size, with ranges of 203–1292 and 75–4249 amino acid residues, respectively, and 12.5–145.0 kDa and 7.3–466.6 kDa in molecular weight, respectively. Their predicted isoelectric points also vary widely from 4.4 to 9.0 and 3.8 to 8.3, respectively. For CPAP1s, this variability is mostly due to the long non-CBD sequence downstream of the CBD that increase the overall size of the protein and, therefore, its overall molecular weight and pI (Jasrapuria et al., 2010). The PMPs vary in the number of CBDs and of linker regions, and consequently vary in size. In contrary, the majority of CPAP3 sequences contain three CBDs and very short linker regions, therefore exhibiting low variability among all the CPAP3s (Jasrapuria et al., 2010).
Table 1.
List of the chitin-binding proteins detected in the genome of M. sexta.
| Gene name/Splice variant | Manduca base Accession# | GenBank Accession# | Protein length (aa) | M.W.a (kDa) | pIa | CBD | Citations | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| CPAP1s | MsCPAP1-A | Msex2.13160 | 203 | 23.1 | 5.7 | 1 | ||
| MsCPAP1-B1 | Msex2.00613 | 193 | 21.4 | 7.6 | 1 | |||
| MsCPAP1-B2 | Msex2.00614 | 730 | 82.3 | 6.4 | 1 | |||
| MsCPAP1-C | Msex2.00381 | 515 | 57.6 | 5.1 | 1 | |||
| MsCPAP1-D | Msex2.03859 | 108 | 12.5 | 5.9 | 1 | |||
| MsCPAP1-F | Msex2.02135 | 124 | 14.2 | 4.4 | 1 | |||
| MsCPAP1-G | Msex2.00269 | 321 | 38.0 | 4.9 | 1 | |||
| MsCPAP1-H | Msex2.02134 | 1037 | 121.3 | 8.8 | 1 | |||
| MsCPAP1-I | Msex2.03076 | 690 | 77.2 | 9.0 | 1 | |||
| MsCPAP1-J | Msex2.03236 | 1292 | 145.0 | 5.8 | 1 | |||
| MsCPAP1-K | Msex2.01703 | 835 | 93.3 | 9.0 | 1 | |||
| MsCPAP1-L | Msex2.09867 | 504 | 49.6 | 5.8 | 1 | |||
| MsCPAP1-M | Msex2.03103 | 731 | 80.1 | 6.1 | 1 | |||
| MsCPAP1-N | Msex2.02137 | 272 | 31.4 | 4.4 | 1 | |||
| MsCPAP1-O | Msex2.08722 | 831 | 94.0 | 7.5 | 1 | |||
| CPAP3s | MsCPAP3-A1 | Msex2.08805 | 237 | 26.3 | 4.9 | 3 | ||
| MsCPAP3-A2 | Msex2.08806 | 240 | 26.8 | 4.8 | 3 | |||
| MsCPAP3-B | Msex2.08808 + Msex2.15015 | 296 | 33.0 | 5.1 | 3 | |||
| MsCPAP3-Ca | Msex2.08810 | 261 | 29.0 | 4.9 | 3 | |||
| MsCPAP3-Cb | Msex2.08810 | 235 | 26.0 | 4.7 | 3 | |||
| MsCPAP3-D1 | Msex2.08807 | 227 | 24.9 | 5.0 | 3 | |||
| MsCPAP3-D2 | Msex2.04890 | 251 | 28.6 | 5.5 | 3 | |||
| MsCPAP3-E1 | Msex2.03293 | 255 | 28.0 | 4.7 | 3 | |||
| MsCPAP3-E2 | Msex2.03294 | 219 | 24.7 | 4.8 | 3 | |||
| MsCPAP3-E3 | Msex2.03295 | 640 | 67.4 | 4.6 | 3 | |||
| MsCPAP3-E4 | Msex2.14886 | 163 | 18.2 | 5.1 | 3 | |||
| PMPs | MsPMP1-A | Msex2.03475 | 657 | 77.8 | 3.8 | 1 | ||
| MsPMP1-B | Msex2.05978 | 154 | 17.3 | 7.1 | 1 | |||
| MsPMP1-C | Msex2.11773 | 4249 | 466.6 | 5.5 | 1 | |||
| MsPMP1-D | Msex2.11956 | 3325 | 365.6 | 5.5 | 1 | |||
| MsPMP1-E | Msex2.15494 | 75 | 7.3 | 4.9 | 1 | |||
| MsPMP1-F | Msex2.05420 | 673 | 73.9 | 8.3 | 1 | |||
| MsPMP2-A | Msex2.04630 | 316 | 34.0 | 4.3 | 2 | |||
| MsPMP2-B | Msex2.08809 | 308 | 33.8 | 5.3 | 2 | |||
| MsPMP2-C | Msex2.09613 | 241 | 26.4 | 4.7 | 2 | |||
| MsPMP3-A | Msex2.04629 | 411 | 43.0 | 4.5 | 3 | |||
| MsPMP3-B | Msex2.05974 | 534 | 52.4 | 4.5 | 3 | |||
| MsPMP4 | Msex2.05986 | 614 | 65.9 | 5.2 | 4 | |||
| MsPMP5 | Msex2.04627 | 543 | 57.3 | 4.5 | 5 | |||
| MsPMP8 | Msex2.05971 | 955 | 105.4 | 5.2 | 8 | |||
| MsPMP9-A | Msex2.00706 | 810 | 90.7 | 4.9 | 9 | |||
| MsPMP9-B | Msex2.05985 | 3467 | 361.1 | 4.1 | 9 | |||
| MsPMP14 | Msex2.02712 | 2076 | 229.1 | 5.0 | 14 | |||
| Chitinases | MsCHT3 | Msex2.04654 | 2271 | 247.6 | 6.1 | 3 | ||
| MsCHT5 | Msex2.08301 | P36362 | 554 | 62.2 | 5.3 | 1 | (Kramer et al., 1993) | |
| MsCHT6 | Msex2.07800 | 3208 | 359.7 | 5.4 | 1 | |||
| MsCHT7 | Msex2.03556 | 996 | 113.4 | 6.3 | 1 | |||
| MsCHT8 | Msex2.10627 | 1181 | 127.9 | 4.8 | 1 | |||
| MsCHT10 | Msex2.04970 | 2898 | 321.9 | 5.9 | 8 | |||
| Chitin deacetylases | MsCDA1 | Msex2.03153 | 539 | 61.5 | 5.0 | 1 | ||
| MsCDA2A | Msex2.03152 | 536 | 60.8 | 5.3 | 1 | |||
| MsCDA2B | Msex2.03152 | 542 | 61.5 | 5.3 | 1 | |||
| MsCDA3 | Msex2.02612 | 525 | 61.0 | 5.8 | 1 | |||
| MsCDA4 | Msex2.02671 | 494 | 56.0 | 5.5 | 1 | |||
| MsCDA5A | Msex2.03714 | 888 | 100.1 | 6.6 | 1 | |||
| MsCDA5B | Msex2.03714 | 888 | 100.3 | 6.8 | 1 | |||
Molecular weight (M.W.) and isoelectric point (pI) were determined using the ExPASy Compute pI/Mw tool available at http://web.expasy.org/compute_pi; CPAP, cuticular proteins analogous to peritrophins; PMP, peritrophic matrix proteins; CBD, chitin binding domain.
Alignment of the CBDs from the proteins in each family of CBPs showed high sequence variability among the three categories, with the six cysteine residues being the only highly conserved motif between all CBPs (Fig. 1). Some aromatic amino acids are also conserved between CBDs in some categories (Supplementary Fig. 1). Such variability is consistent with the previous reports from other species, such as D. melanogaster (Behr and Hoch, 2005), Lucilia cuprina (Elvin et al., 1996; Tellam et al., 2003), Mamestra configurata (Shi et al., 2004), T. castaneum (Jasrapuria et al., 2010) and Trichoplusia ni (Wang and Granados, 1997; Wang et al., 2004). This consensus sequence motif of the six cysteine residues of the ChtBD2 domain found in insects has been defined as CX15–17CX5–6CX9CX12CX6–7C (Tellam et al., 1999). It was modified recently with additional data from a genome-wide search of CBDs from T. castaneum to a new consensus, CX11–24CX5CX9–14CX12–16CX6–8C (Jasrapuria et al., 2010). With more ChtBD2 sequences identified from M. sexta CBPs in this study, this ChtBD2 consensus is updated to CX11–30CX5–6CX9–24CX12–17CX6–12C, to include most of the CBDs described in insects so far. Therefore, all differences observed between this consensus and the previous one are due to specific characteristics of M. sexta CBPs. It is to be noted that the CBDs from the enzymes do not differ significantly from those of other CBPs, with the exception of the first CBD in MsCHT3 which has an extended length between the 3rd and 4th cysteine residues (Supplementary Fig. 1).
Fig. 1.

Consensus of conserved cysteines (C) and spacings (×) of the ChtBD2 chitin-binding domain (CBD) for each category of chitin-binding protein (CBP). For each category of CBP, the numbers indicate the lowest and highest number of amino acids (×) found between the conserved adjacent cysteines (C) of the CBD. An overall consensus, taking into account the variability of spacing observed in all the ChtBD2-containing proteins is also shown. The alignments of all the CBDs sequences for each category of CBP are shown in. Supplementary Fig. 1.
3.2. Gene expression patterns
Hierarchical clustering analysis of CBPs gene expression patterns in M. sexta shows that most of the PMPs were clustered together, indicating their shared gene expression in the midgut at nearly all larval instars (Fig. 2). This pattern of expression is consistent with previous observations, notably in L cuprina (Tellam et al., 2003), R. proxilus (Ribeiro et al., 2014), T castaneum (Jasrapuria et al., 2010) and T. ni (Wang et al., 2004), and with the definition of the PMP family (Jasrapuria et al., 2010; Tellam et al., 1999). Expression of some PMP genes was also detected in Malpighian tubules and fat body. However, the tissue origin for the expression of these PMP genes cannot be absolutely defined (e.g. Malpighian tubule or fat body cells vs. attached tracheal cells). The function of these PMPs expressed in the Malpighian tubules and fat body needs to be understood. A few PMPs had divergent expression profiles and these gene showed low global expression (MsPMP1-C, MsPMP1-F, MsPMP2-B, MsPMP14). No clear clustering was observed for CPAPs and chitin metabolism enzymes. Instead, highly diverse gene expression patterns were observed for CPAPs and chitin metabolism enzymes. These patterns are described in details by Tetreau et al. (Tetreau et al., in review) and Dittmer et al. (Dittmer et al., in revision).
Fig. 2.
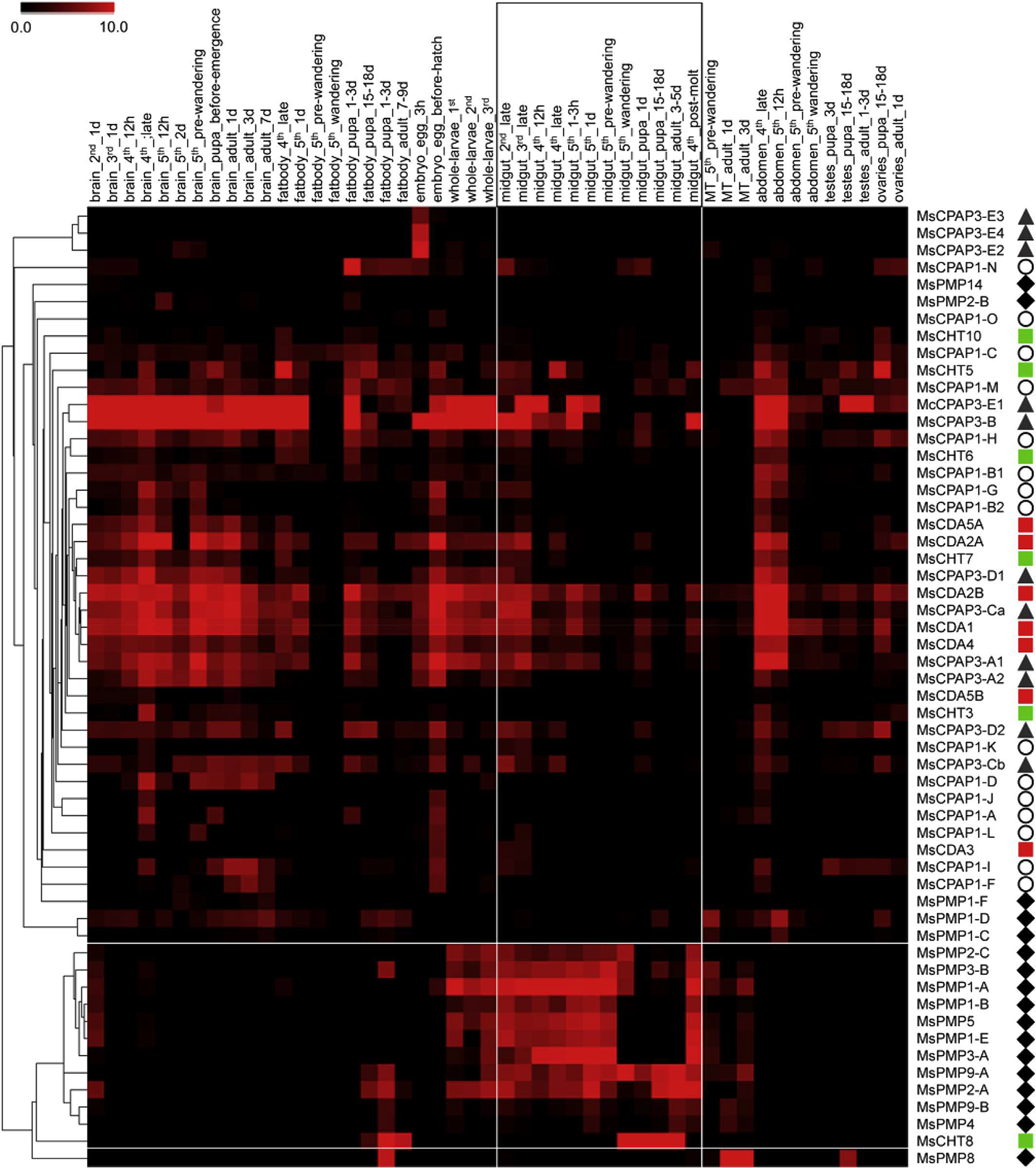
Heatmap of expression and hierarchical clustering analysis of the 56 genes (right of figure) encoding proteins containing chitin-binding domains (CBDs) in different tissues and at different developmental stages (48 RNAseq libraries, top of the figure) in M. sexta. Each library name contains the tissue (brain, fat body, embryo, whole larvae, midgut, malpighian tubules, abdomen, testes and ovaries), the developmental stage (egg, 2nd, 3rd, 4th and 5th larval instars, pupa and adult) and the age within the developmental stage. A pictogram has been associated with each category of CBP: CPAP1s (white circles), CPAP3s (gray triangles), PMPs (dark diamonds), chitinases (green square) and chitin deacetylases (red squares). The midgut specific libraries and the genes specifically expressed in the midgut, as revealed by the clustering analysis, have been highlighted by white lines. The color range representing gene expression goes from black (no expression) to red (high expression). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Phylogenetic analysis of chitin-binding domains
3.3.1. Phylogenetic analysis of CPAP1s
CPAP1 proteins have been extensively studied only in the coleopteran T. castaneum until now (Jasrapuria et al., 2010, 2012). It was of interest to determine whether a lepidopteran insect such as M. sexta had a similar assortment of CPAP1 genes. A total of 15 genes encoding CPAP1 proteins were identified in the M. sexta genome and they were clearly clustered into different groups in the phylogenetic analysis that includes representative members from different insect orders (Fig. 3). Ten of these proteins appeared to be homologous to the CPAP1 proteins of T. castaneum indicating that CPAP1 proteins are not restricted only to the beetle. It is to be noted that the group B initially described in T. castaneum (Jasrapuria et al., 2010) has been divided into two distinct new subgroups (groups CPAP1-B1 and CPAP1-B2), each including one CPAP1 representative from M. sexta and T. castaneum. In addition, five novel proteins were identified and could not be placed as orthologs of the ten groups previously described by Jasrapuria et al. (2010). A search of the insect genome databases using these five M. sexta CPAP1 proteins identified presumptive orthologs in several insect genomes, including T. castaneum, and D. melanogaster. Similar results were independently obtained recently (Ioannidou et al., 2014). Therefore, five new groups including these proteins (named CPAP1-K to CPAP1-O) were created. The clusters in the CPAP1 tree are supported by high bootstrap values, which is consistent with previous observations from T castaneum (Jasrapuria et al., 2010). A previous analysis of CPAPs suggested that Lepidoptera might have a limited number or lack CBPs of the CPAP1 family (Jasrapuria et al., 2012). In the M. sexta genome, a CBP protein in each CPAP1 group, with the exception of group E, was found (Fig. 3). Therefore, Lepidoptera have genes coding for diverse CPAP1 proteins and these genes are expressed, as determined by RNA-seq. However, to verily whether these proteins are indeed cuticular proteins will require validation of the presence of these proteins in the cuticles.
Fig. 3.
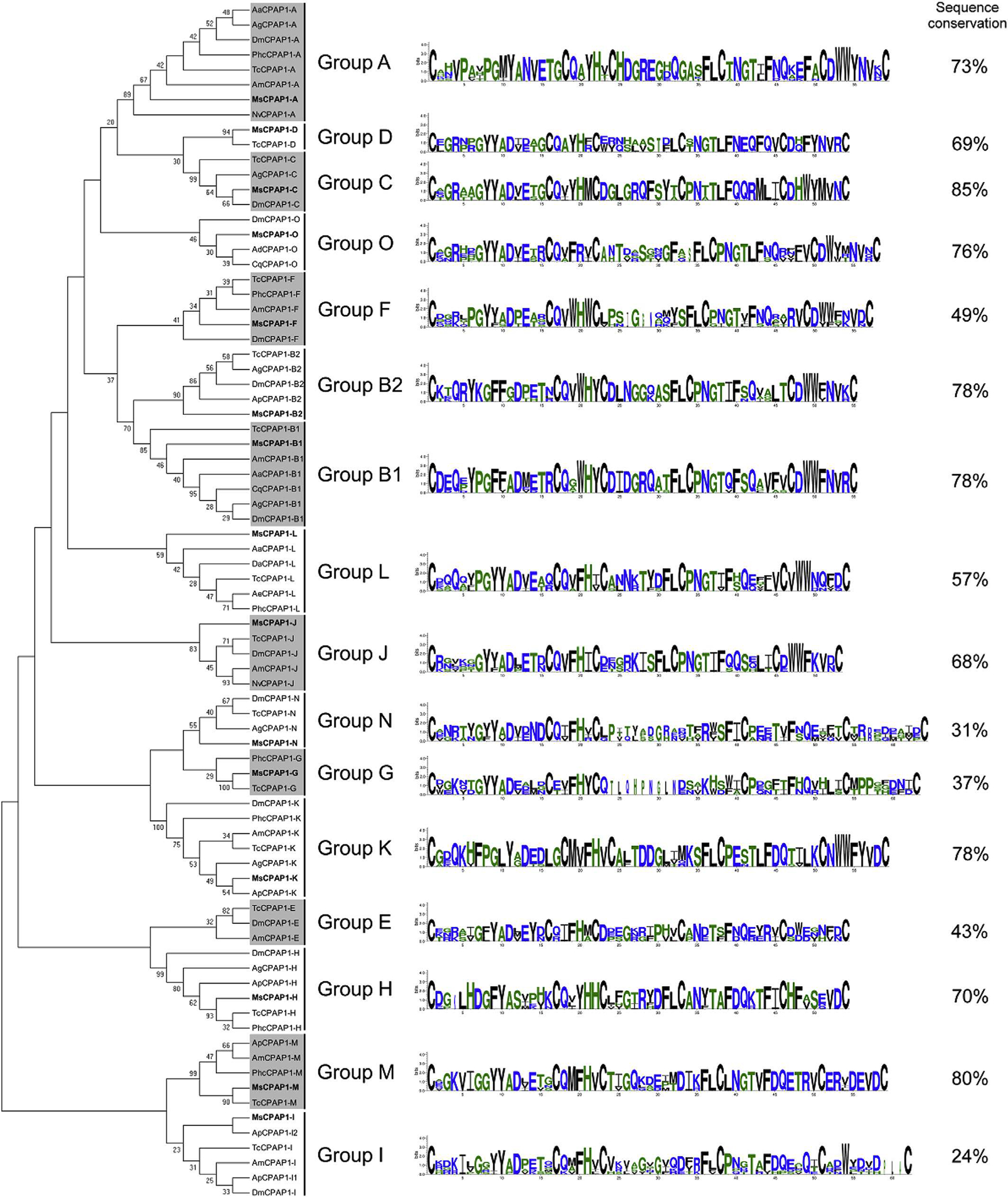
Phylogenetic analysis of the chitin-binding domain of CPAP1s from thirteen different insect species; Acromyrmex echinatior (Ae), Acyrthosiphon pisum (Ap), Aedes aegypti (Aa), Anopheles darling (Ad), Anopheles gambiae (Ag), Apis mellifem (Am), Culex quinquefasciatus (Cq), Drosophila ananassae (Da), Drosophila melanogaster (Dm), M. sexto (Ms), Nasonia vitripennis (Np), Pedicuius humanus corporis (Phc), Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. CPAP1 s from M. sexta are indicated in bold. CPAP1s are grouped into sixteen different groups; the sequence logos of the chitin-binding domain and the percentage of sequence conservation (identical amino adds between all sequences) are shown on the right for each group.
Although CPAP1s were clustered into distinct groups, the sequence conservation between proteins within the same group varies from one group to another. While half of the groups (groups A, B1, B2, C, H, K, M and O) exhibited a sequence conservation higher than 70% (with a maximum of 85% for group C), some groups exhibited a very low sequence conservation, such as groups I and N with 24% and 31% of sequence conservation, respectively (Fig. 3). RNAi experiments performed in T. castaneum revealed that CPAP1-C, CPAP1-H and CPAP1-J were important for the survival of this insect. Knock-down of these genes affected pupal-to-adult morphogenesis (Jasrapuria et al., 2012). Given the level of sequence conservation of CPAP1s in the groups C, H and J (85%, 70% and 68%, respectively), we predict that these CPAP1s may be involved in similar biological functions in M. sexta and other Lepidoptera.
3.3.2. Phylogenetic analysis of CPAP3s
The CPAP3 family is highly conserved in term of domain organization and sequence similarity and it is present in all insect orders examined (Jasrapuria et al., 2012; Willis et al., 2012). Seven phylogenetic groups were observed by analyzing the full sequences of CPAP3s from different species belonging to several insect orders (Fig. 4). The grouping is supported by high bootstrap values and is consistent with the groups described in T. castaneum (Jasrapuria et al., 2010). Analyses of the CPAP3s from 10 species either in full length or in individual domains showed the same grouping, with only slight differences from one CBD to another (Supplementary Fig. 2). Phylogenetic analysis of all the CBDs from CPAP3s showed a highly structured clustering of the CBDs (Fig. 5). First, the CBD sequences were clustered in three different groups corresponding to the three CBD domains (CBD1, CBD2 and CBD3), except for the CBD2s in CPAP3-D2 proteins that were clustered with CBD1s (Fig. 5). Among the three CBDs in CPAP3s, CBD1s are more closely related to CBD2s than to CBD3s. Moreover, relatively high sequence conservation is observed for CBDs from the same rank in each group (45–92% sequence identity), except for group E (14–23% sequence identity) (Supplementary Fig. 3). Secondly, CPAP3s have the same CBD structure with the 3 tandem CBDs in the same order, indicating that the multiple members (seven) of CPAP3 family in insects were derived from gene duplications. Finally, CPAP3 proteins are more closely related to CPAP3s within the same groups than to other CPAP3s from the same insect species. Therefore, CPAP3s arose and evolved to become a multiple gene family prior to the divergence of ancestral insect lineages to the current different orders.
Fig. 4.
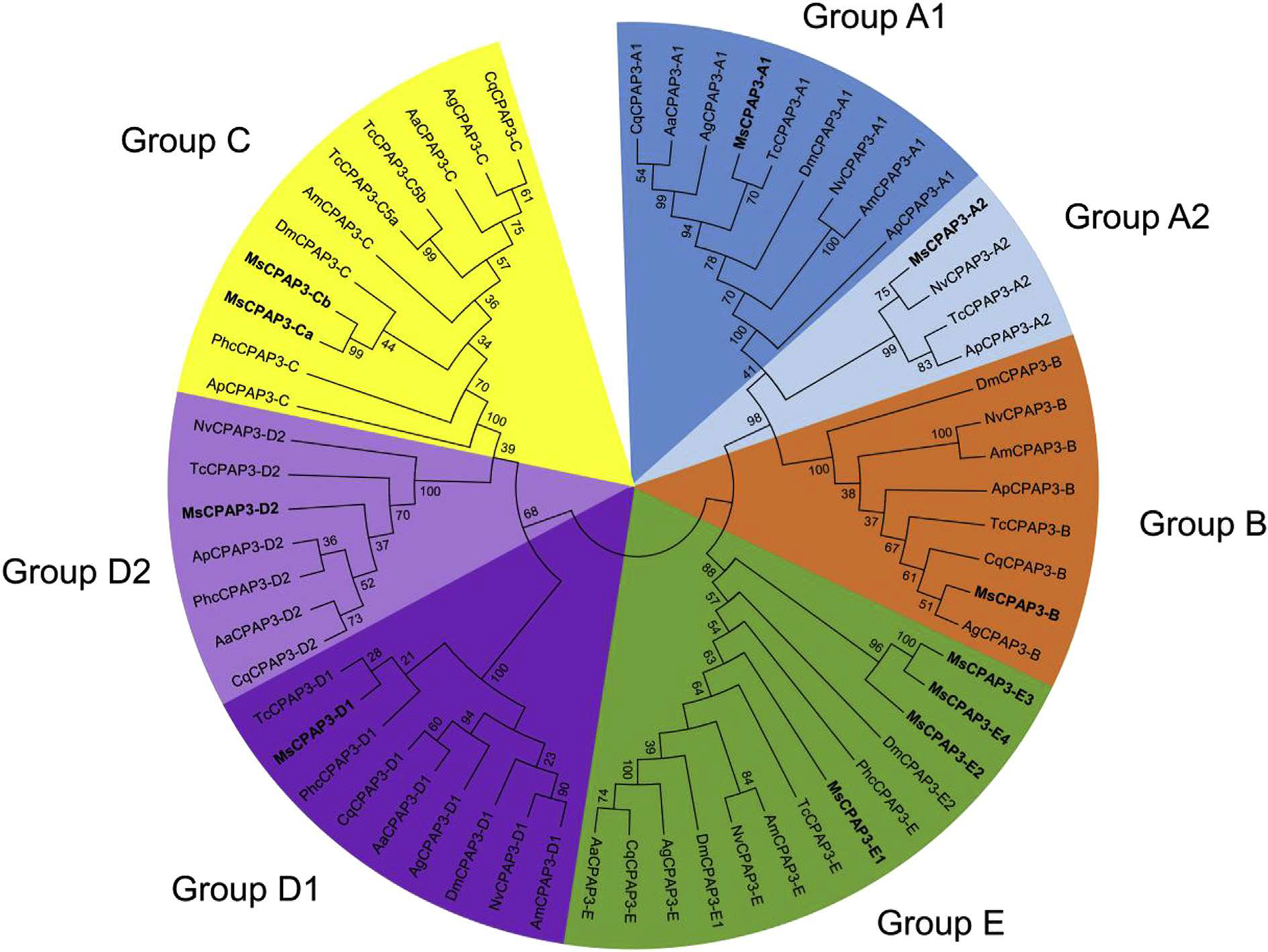
Phylogenetic analysis of the full protein sequences of CPAP3s from ten different insect species; Acyrthosiphun pisum (Ap), Aedes aegypti (Aa), Anopheles gambiae (Ag), Apis mellijera (Am), Culex quinquefasdatus (Cq), Drosophila melanogaster (Dm), M. sexta (Ms), Nasonia vitripennis (Np), Pediculus humanus corporis (Phc), Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. CPAP3s are grouped into seven different groups; group A1 (dark blue), groups A2 (light blue), group B (orange), group C (yellow), group D1 (dark purple), group D2 (light purple) and group E (green). CPAP3s from M. sexta are indicated in bold. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
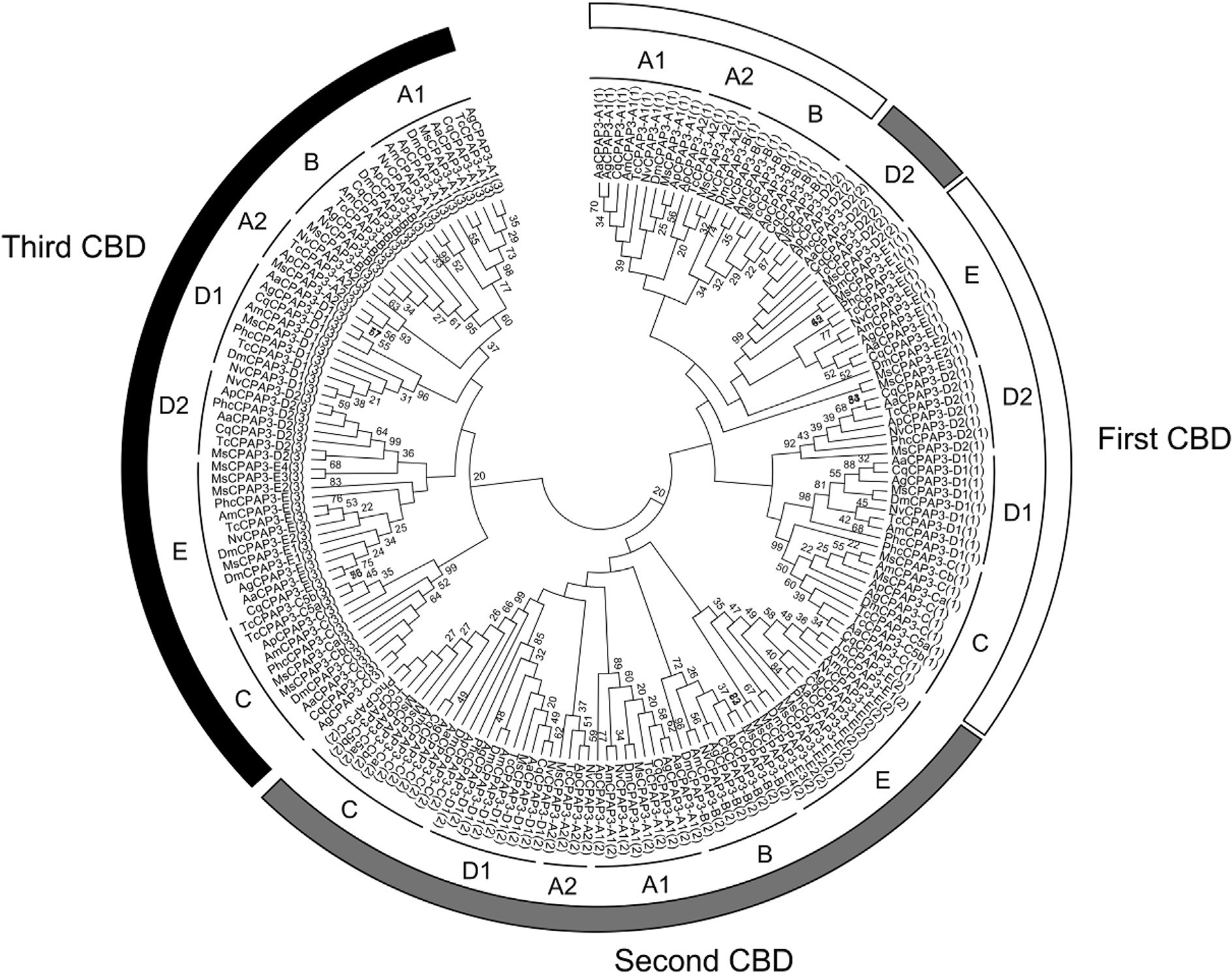
Phylogenetic analysis of the three chitin-binding domains (CBDs) of CPAP3s from ten different insect species; Acyrthosiphon pisum (Ap), Aedes aegypti (Aa), Anopheles gambiae (Ag), Apis mellifem (Am), Culex quinquefasciatus (Cq), Drosophila melanogaster (Dm), M. sexta (Ms), Nasonia vitripennis (Np), Pediculus humanus corporis (Phc), Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. The seven different groups of CPAP3s are indicated (from A to E). The rank of the CBD is also indicated by a number (from 1 to 3) in brackets at the end of the CPAP3 name and by a color (first CBD, white; second CBD, gray; third CBD, black).
The function of CPAPs has not been well studied in Lepidoptera, but it has been studied in a few cases in D. melanogaster and more extensively T. castaneum (Jasrapuria et al., 2012; Petkau et al., 2012). In D. melanogaster, disruption of obstructor-A (named DmCPAP3-A1 in this study) resulted in severe defects during cuticle molting, wound protection and larval growth control, indicating that CPAP3-A1 is involved in the protection of chitin from early degradation and in the maintenance of the proper size, structure and function of the cuticle (Petkau et al., 2012). Moreover, the involvement of Obst-A and of “gasp” (gene analogous to small peritrophins) proteins, which are CPAP3 proteins from group A, in the expansion and preservation of the integrity of airway tube was also evidenced in obst-A and gasp null mutant lines of Drosophila (Tiklova et al., 2013). In T. castaneum, RNAi of individual CPAP3 genes led to different phenotypes: inhibition of CPAP3-A1 resulted in fat body depletion and defecation problems leading to adult death; CPAP3-B knock-down resulted in alteration of leg articulations inducing a stiff and uncoordinated gait; CPAP3-C RNAi was lethal resulting from failure of pupal-to-adult molt; CPAP3-D1 and -D2 inhibition affected the morphology of elytral cuticle, while the knock-down of CPAP3-A2 and CPAP3-E induced no noticeable phenotype modification (Jasrapuria et al., 2012). These observations indicate that many CPAP3s are functionally non-redundant and play essential roles in metamorphosis, locomotion and in structural integrity and dynamic maintenance of cuticle in insects. Given the high sequence conservation of among subgroups of CPAP3s from different insect species, it is expected that the distinctive functions of each subgroup of CPAP3 orthologs may be conserved in insects.
3.3.3. Phylogenetic analysis of PMPs
A total of 67 CBDs from 17 different PMPs were identified in M. sexta (table 1). Phylogenetic analysis of PMP domain sequences from M. sexta and T. castaneum (Jasrapuria et al., 2010) did not show any clear clustering by species or by phylogenetic groups based on sequence homology (Fig. 6). The same pattern is observed even when PMPs from additional species were included in the analysis (Supplementary Fig. 4). Many of the CBDs from the same protein tend to cluster together, suggesting that contrary to what has been observed in CPAP3s, the events leading to an increase in the number of CBDs in PMPs occurred after the insect species diversification, although PMP domains arose prior to the divergence of insect orders.
Fig. 6.
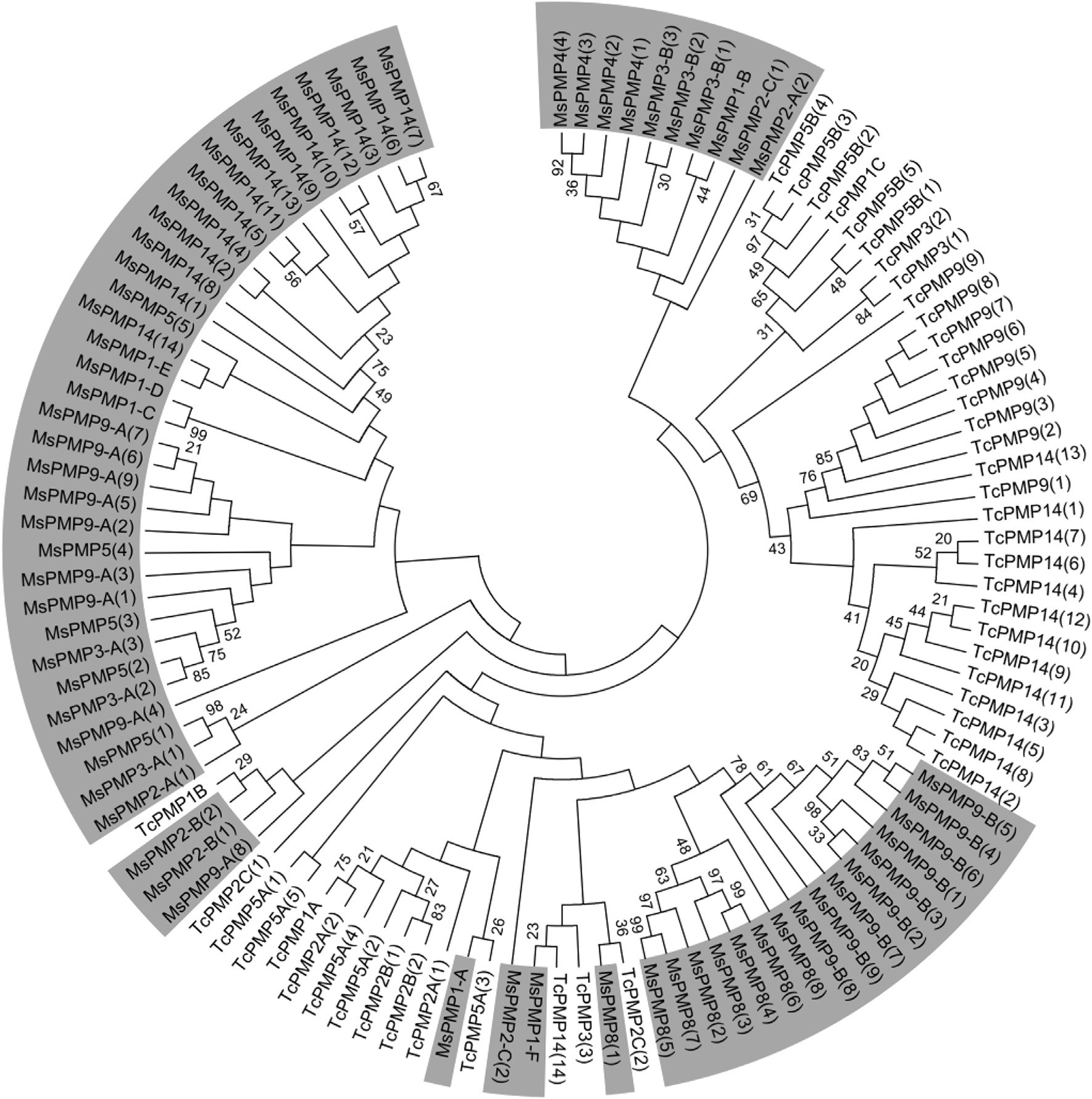
Phylogenetic analysis of the chitin-binding domains (CBDs) of PMPs from M. sexta (Ms) and from Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. A tree including additional species is available in Supplementary Figure PMP Phylo. The rank of the CBD in the PMP is indicated by a number (from 1 to 14) in brackets at the end of the PMP name. PMPs from M. sexta are highlighted in gray.
PMPs are primarily expressed in the midgut (Jasrapuria et al., 2010), and their functions have been proposed to bind and cross-link chitin fibrils to form the PM structure. In a recent study, knocking down 2 of the 11 PMP genes from T. castaneum by RNAi (TcPMP3 and TcPMP5-B) caused abnormal larval growth and molting, and eventually death (Agrawal et al., 2014). Inhibition of the other 9 PMP genes did not show any noticeable abnormal phenotype, suggesting that they may have redundant functions and that they are not mandatory for insect development and survival. As the number and domain organization of PMPs appear to be species-specific, their functions may vary from one species to another and may also depend on their feeding habits. The role of PMPs in the regulation of PM integrity and functions requires to be investigated in multiple species in order to acquire a full understanding of the functions of this family of proteins.
3.3.4. Phylogenetic analysis of CBD sequences from chitinases (CHTs)
In M. sexta, 11 CHTs were identified and classified into ten different groups based on the sequences of their catalytic domain (Tetreau et al., 2015). Among the CHTs, six groups contained at least one ChtBD2-type CBD. Phylogenetic analysis revealed that the clustering of the CHTs by using CBD sequences was identical to the clustering obtained with catalytic domain sequences from the same CHTs in groups I, VI and X (Fig. 7). This observation suggests that the CBD became associated with these active CHTs before the divergence of insect orders. AH CHTs from the group X from different insect orders contained three CBDs. The first, second and third CBDs of group X CHTs form three clearly distinct clades (Fig. 7). This suggests that the CBD multiplication event that led to the appearance of group X CHTs is an ancient event that preceded the divergence of insect orders.
Fig. 7.
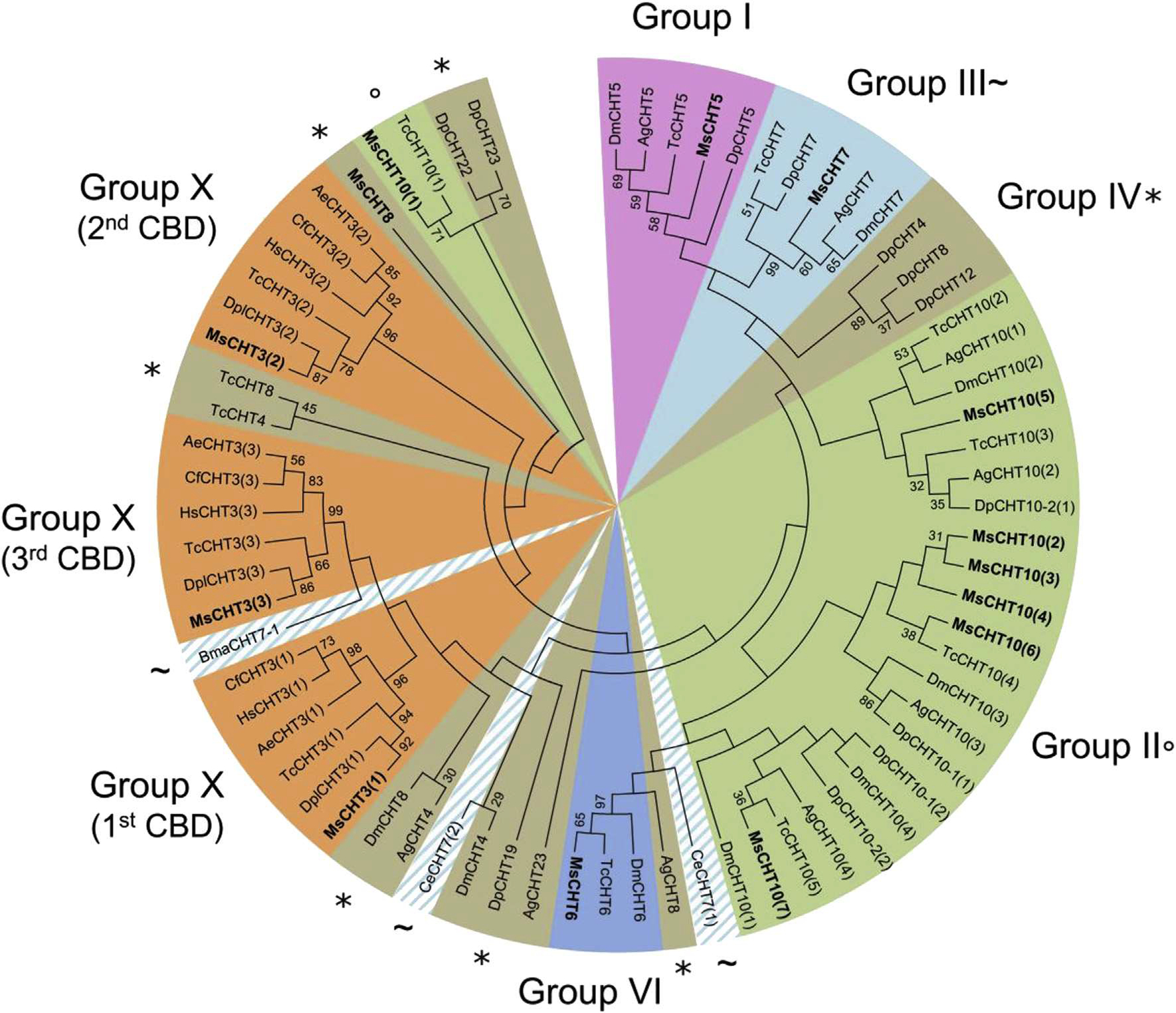
Phylogenetic analysis of all chitin-binding domains (CBDs) of chitinases from eleven different insect species: Acromyrmex echinatior (Ae), Anopheles gambiae (Ag), Brugia malayi (Bma), Caenorhabditis elegans (Ce), Camponotus floridanus (Cf), Danaus plexippus (Dpl), Daphnia pulex (Dp), Drosophila melanogaster (Dm), Harpegnathos saltator (Hs), Manduca sexta (Ms) and Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. CHTs containing a CBD are grouped into 6 different groups out of the 10 described for CHTs (Tetreau et al., 2015): group I (light purple), group II (green; °), group III (light blue; ~), group IV (brown; *), group VI (dark blue) or group X (orange). As CHT7 chitinases from C. elegans and B. malayi (indicated by a ~) have a divergent domain organization from other group III chitinases (Tetreau et al., 2015), they are highlighted by a hatched light blue area. CHTs from M. sexta are indicated in bold. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Group III CBDs were represented in the tree as a main block containing CHT7s from Diptera, Coleoptera, Crustacea and Lepidoptera but not the CHT7 from Nematoda (BmaCHT7–1 and CeCHT7). Analysis of domain organization and sequences of catalytic domains of CHT7s also revealed a separation between CHT7s from Nematoda and the ones from other species (Tetreau et al., 2015). This supports the hypothesis of an ancestral domain organization and sequences of catalytic domains (Tetreau et al., 2015) and CBDs (present study) in CHT7s in Nematoda, which has further evolved and diversified in groups III in Arthropoda.
Group II includes CHTlOs that have 4–5 catalytic domains and 4–8 CBDs (Arakane and Muthukrishnan, 2010; Tetreau et al., 2015). The CBDs from the CHT10s were all clustered together, except for the first CBD from M. sexta and T. castaneum that form a clear distinct group (Fig. 7).
The CBDs associated with group IV CHTs were scattered widely across the tree and no clear clustering could be observed. This group is the largest group of CHTs but have the simplest structure based on their domain organization (Arakane and Muthukrishnan, 2010) and most group IV CHTs do not contain any CBD or C-terminal extensions. This suggests that CBDs in this group are rare and that they were acquired in these CHTs more recently, after the diversification of the CHT family, probably from multiple CBD duplication events from different origins.
Altogether, these analyses suggest that CBDs evolved along with the catalytic domain in CHTs in most phylogenetic groups before the divergence of the CHT families, except for group IV in which CBDs were acquired more recently and were only found in few proteins from some insect species.
3.3.5. Phylogenetic analysis of CBD sequences from chitin deacetylases (CDAs)
CDAs are generally classified into five distinct phylogenetic groups based on catalytic domain sequences homology, of which four contain CDAs with a single CBD in their N-terminal sequence (Dixit et al., 2008). Phylogenetic analysis of CBDs from CDAs showed the same grouping of the CDAs as that obtained using the catalytic domain sequences, with four distinct groups (groups I to IV; Fig. 8) (Tetreau et al., 2015). This suggests that the CBDs became associated with the catalytic domain before the emergence and diversification of insect orders, as it is observed for most CHTs.
Fig. 8.
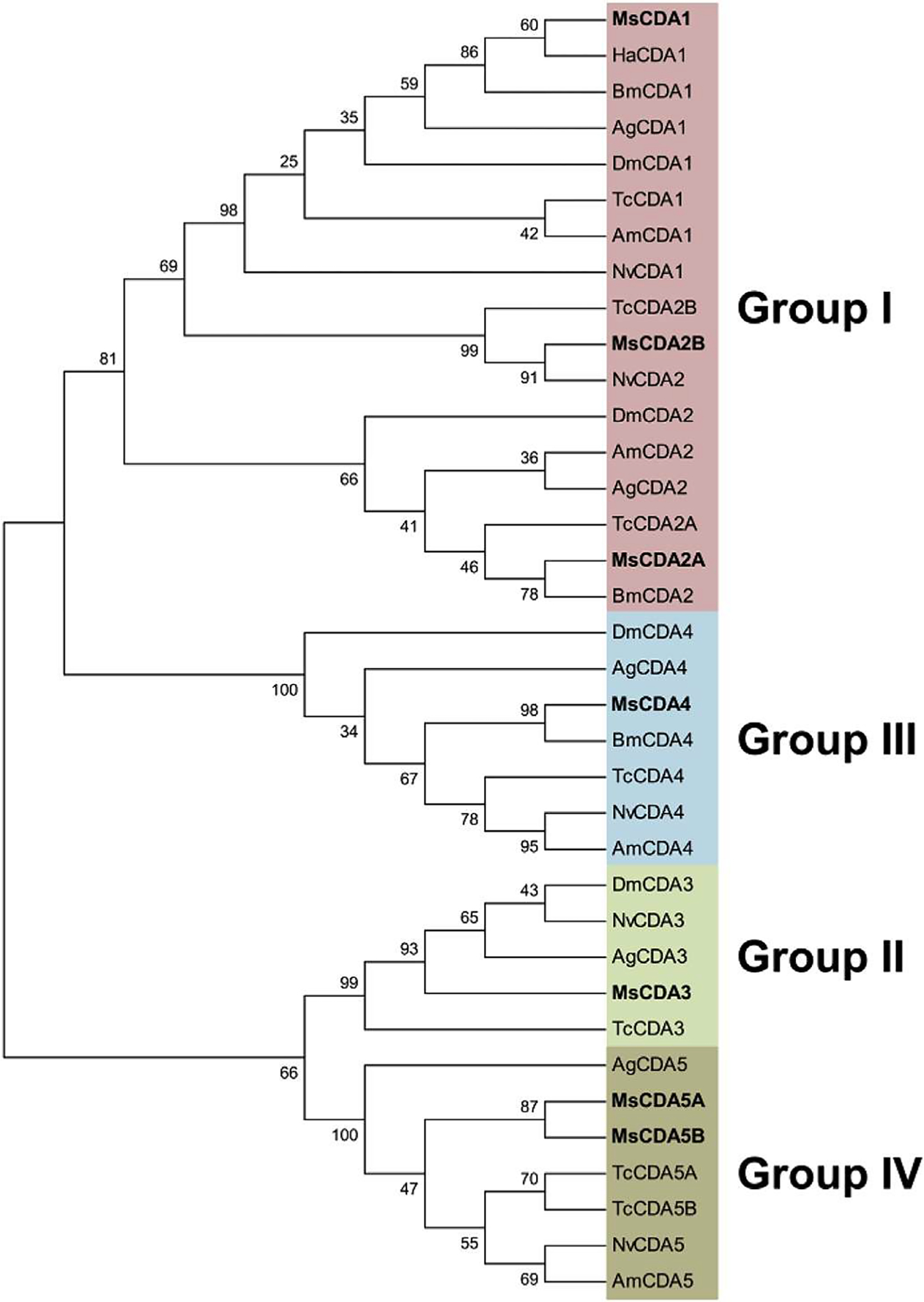
Phylogenetic analysis of the chitin-binding domain (CBD] of chitin deacetylases (CDAs) from eight different insect species; Anopheles gambiae (Ag), Apis mellifem (Am), Bombyx mori (Bm), Drosophila melanogaster (Dm), Helicoverpa armigera (Ha), Manduca sexta (Ms), Nasonia vitripennis (Nv) and Tribolium castaneum (Tc). The accession numbers of all the proteins used are listed in Supplementary Table 1. CDAs containing a CBD are grouped into 4 different groups out of the 5 described for CDAs (Dixit et al., 2008): group I (pink), group II (green), group III (light blue) or group IV (brown). CDAs from M. sexta are indicated in bold. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Origin of CBDs in CBPs
CBPs are the most important proteinaceous components for the structures of cuticle and PM in insects (Terra and Ferreira, 2005; Willis et al., 2005). While it is generally accepted that a unique evolutionary event allowed the appearance of the cuticle, as an exaptation allowing the Ecdysozoa to colonize the continents (Labandeira, 2005), the origin of the PM, notably in insects, appears to be more complex and remains unclear. It has been proposed that ancestral insects had a substance containing mucin (peritrophic gel) that covered the gut epithelium, like most vertebrate animals, and that the PM was derived from the mucus (Terra, 2001). According to this model, peritrophins evolved from mucins by acquiring CBDs to become the unique insect intestinal mucins which contain both mucin domains and CBDs (Wang and Granados, 1997), and later some of them lost their mucin-like domains to become the current peritrophins (Terra and Ferreira, 2005). The concomitant secretion of chitin and peritrophins by the midgut epithelium permits the formation of the chitin-protein network – the structural foundation of insect PM (Terra, 2001).
3.4.1. Origin and evolution of CBDs in CPAPs
The genome-wide search and phylogenetic analysis of CBPs and CBDs from M. sexta and other species indicate that the cuticular CBPs (i.e. CPAP1s and CPAP3s) are highly conserved across several orders of insects (Ioannidou et al., 2014; Jasrapuria et al., 2010; Willis et al., 2012). Functional studies of multiple CPAPs in T. castaneum by RNAi have shown that CPAPs play essential non-redundant roles in T. castaneum (Jasrapuria et al., 2012). Similarly, in D. melanogaster some CPAP3s have been shown to be important for larval survival and/or development (Behr and Hoch, 2005; Petkau et al., 2012). While nematodes and the sea urchin do have proteins with ChtBD2 domains, they appear to be related to the peritrophins and no orthologs to CPAPs could be found in these species. The appearance of many CPAP1 and CPAP3 proteins in the crustacean Daphnia pulex indicates that the expansion of the two families of CPAPs may have been selected during evolution to perform conserved, but non-redundant, functions in arthropods. The conservation of multiple members of CPAP1 and CPAP3 families and their unique subfamily sequences is consistent with the hypothesis of the ancient appearance of the cuticle, which allowed protecting arthropods from their environment and predators, and it also suggests that the current assortment of CPAP1 and CPAP3 families was fixed in arthropod genomes prior to the evolutionary radiation of insects.
3.4.2. Origin and evolution of CBDs in PMPs
In contrast to CPAPs, the number of PMPs in a species and the number of CBDs in PMPs vary drastically from one species to another. For example, the CBDs in PMPs from M. sexta and T. castaneum do not show clustering between species but rather cluster with other repeats in the same protein or among repeats from closely linked genes (e.g. MsPMP9 and MsPMP14). Contrary to CPAP3s, whose domain replication and fixation of their consensus sequences occurred before the divergence of insect from other arthropoda, the multiplication of CBDs in PMPs is predicted to have occurred after speciation. The PM and ancestral versions of PMPs were supposedly acquired by insects and some other invertebrates before their evolutionary radiation (Terra, 2001). Genes encoding proteins with multiple peritrophin-A domains (e.g. XP_001895704) and some with interspersed mucin domains (e.g. ABC65811) can be found in nematoda and in crustacean genomes. However, the divergence of the PMP family among different insect orders suggests that the diversification of the members of PMP family involved in the chitin-protein complex of the PM was a more recent event and may have continued after insect speciation.
3.4.3. Origin and evolution of CBDs in CPAPs and PMPs – description of the model
The phylogenetic analysis of the sequence similarities among CPAP1s, CPAP3s and PMPs provided additional insights into evolution of CBDs (Fig. 9). CPAP1s formed a clade clearly separated from CPAP3s and PMPs in the phylogenetic tree. The clear clustering of CPAP1s as a separate group is consistent with the observation that the sequences of members of subgroups of CPAP1s are highly conserved among different insect species. However, the clear dichotomy reported between CBDs of CPAP3s and PMPs in T. castaneum (Jasrapuria et al., 2010) was not observed in M. sexta. Instead, the CBDs from PMPs and CPAP3s were mixed together in the same branches in M sexta, when either only the first CBD (Fig. 9) or all the CBDs (Supplementary Fig. 5) were used for the phylogenetic analysis. The same results were obtained when we included CBPs from additional species (Supplementary Fig. 6). This observation suggests that the CBDs in PMPs may have originated from CPAP3 CBDs, followed by duplication and transposition into a mucin-like gene or into gut-specific genes leading to ancestral PMPs. The multiplication of CBDs, which exhibit different levels of sequence divergence between PMPs from the same species and between proteins from different insect species, appear to be a species-specific event, suggesting a recent gene family expansion. The large number of CBDs in PMPs specific to insect species may be an adaptive evolution of the protein family to the protease-rich gut environment (Wang et al., 2004). Feeding pattern modification is known to be a powerful evolutionary force leading to speciation in insects (Bass et al., 2013; Cates, 1980; Simonato et al., 2013). The variations in the number and sequence of CBDs in PMPs may also be an adaptive evolution of this family of proteins to food preferences, which differs from one insect species to another. The fact that only a limited number of PMPs have been shown to be essential for normal PM functional integrity and insect survival (Agrawal et al., 2014) supports the notion that a more recent expansion of the PMP family allowed adaptation of insects to diverse hosts. Additional genome sequence information from more insect species and more studies on CBPs functions are needed to better understand the roles of PMPs in insects.
Fig. 9.
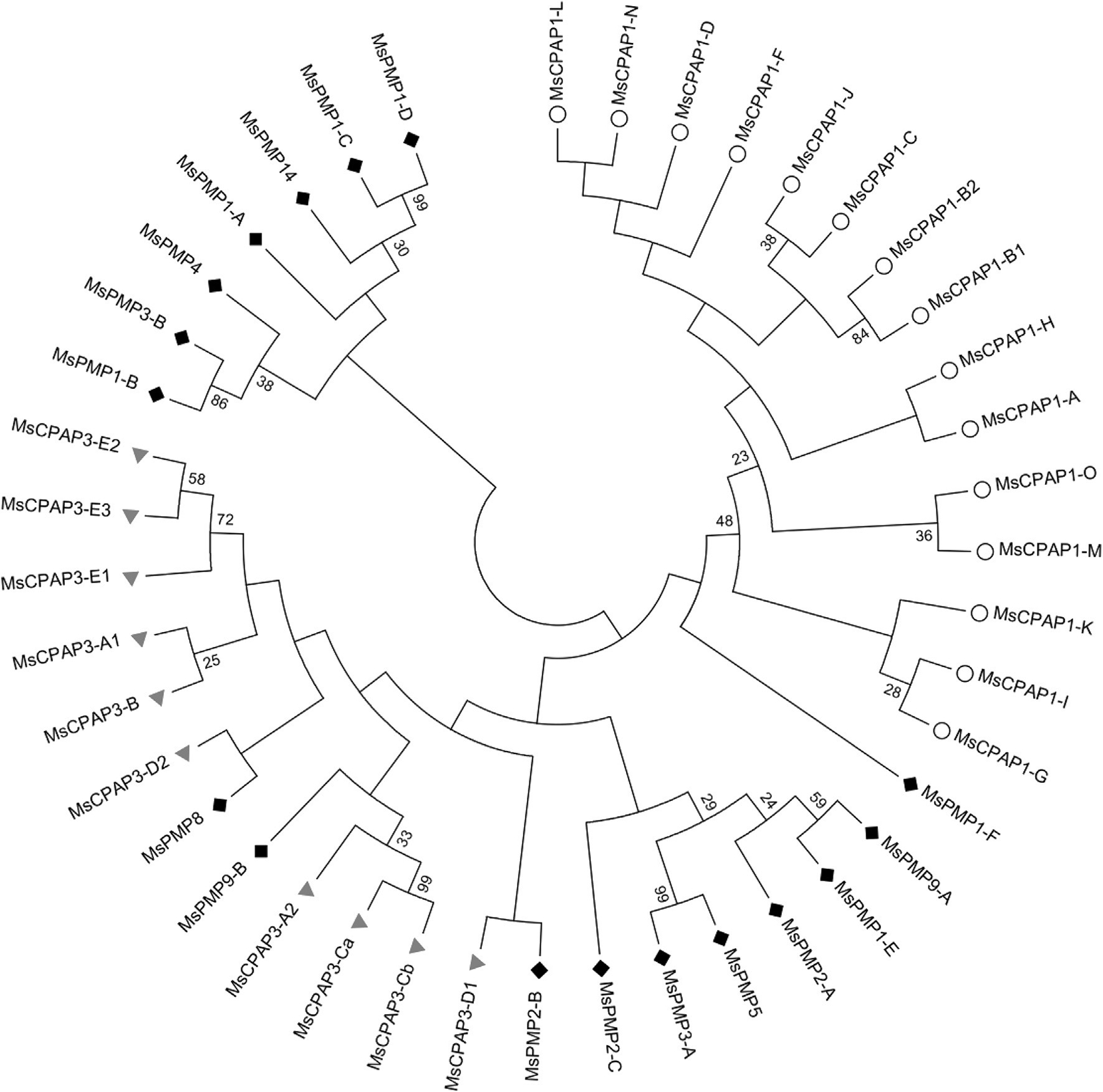
Phylogenetic analysis of the chitin-binding domain (CBD) of CPAP1s (white circle) and of the first CBD of CPAP3s (gray triangle) and PMPs (dark diamond) from Manduca sexta.
3.4.4. Origin and evolution of CBDs in chitin metabolism enzymes
The patterns of clustering of CHTs and CDAs from phylogenetic analysis were the same using either the CBD sequences or the catalytic domains (except for group IV CHTs). This suggests that the CBDs in the CHTs and CDAs were associated with the corresponding CHT or CDA catalytic domains before the divergence of the insect orders, so that CBDs evolved along with the catalytic domains in the same proteins. The CBDs from CHTs and CDAs do not segregate into two separate groups as one would expect for the two functionally completely different enzyme families (Fig. 10A). These results indicate that ancient CHTs and CDAs were devoid of CBDs and that the latter was acquired later during evolution presumably from a common precursor with this domain. Consistent with this notion is that both classes of enzymes have representatives with and without the CBDs. The CBD domains of the CPAP1s are clearly separated from those of the CPAP3s, PMPs and ChtBD2-containing enzymes, while CBDs from CPAP3s, PMPs and ChtBD2-containing enzymes are interspersed among different clades in the phylogenetic tree. These observations are clearly shown by analyses of the proteins from M. sexta alone (Fig. 10B) and with proteins from 23 other species (Supplementary Fig. 6). Even though the CBDs from PMPs, CPAP3s and enzymes form a large clade distinct from the CPAP1 CBDs, whether all of these CBDs came from a single precursor such as one of the CBDs of CPAP3 is yet to be determined.
Fig. 10.
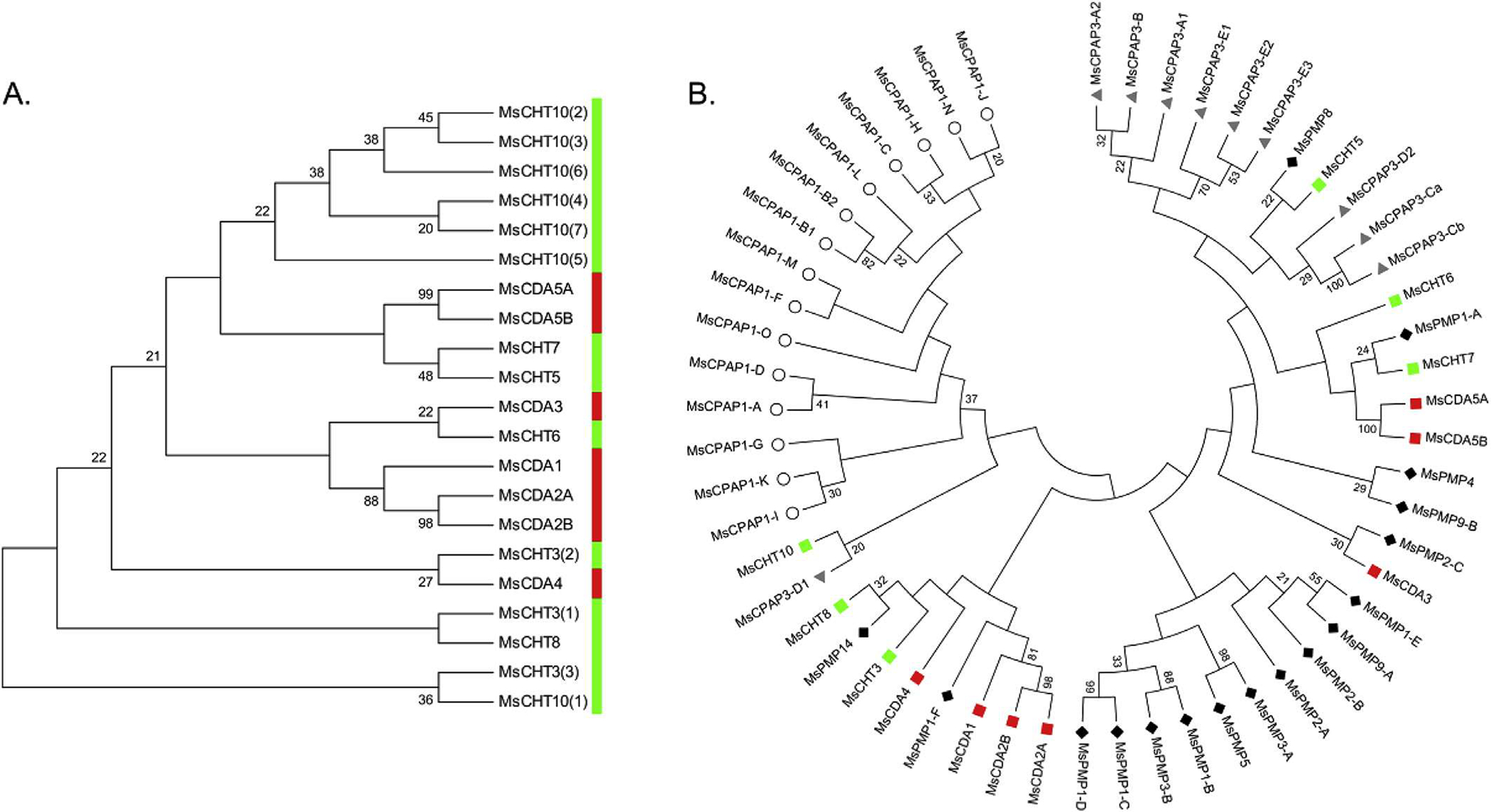
A. Phylogenetic analysis of the chitin-binding domains (CBDs) of chitinases (green) and chitin deacetylases (red) from M. sexta. The numbers in brackets at the end of the enzyme name indicate the rank of the CBD in proteins with multiple repeats. B. Phylogenetic analysis of the first chitin-binding domain of CPAP1s (white circle), CPAP3s (gray triangle), PMPs (dark diamond), chitinases (green square) and chitin deacetylases (red square) from Manduca sexta. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
Proteins binding to chitin are key structural components of the cuticle and the PM in insects and are dynamically involved in formation of chitin-containing structures and modulation of their functions. A genome-wide search and analysis of chitin-binding proteins in M. sexta and phylogenetic analyses of CBDs from different arthropods have provided clues concerning the evolution of chitin-binding proteins in a Lepidopteran genome as well as in insects of different orders. A total of 53 genes encoding 56 ChtBD2-containing CBPs or enzymes were identified in the M. sexta genome, including 15 CPAP1s, 11 CPAP3s, 17 PMPs, 6 CHTs and 7 CDAs. RNA-seq analysis provided a global view of the expression patterns of these CBPs in M. sexta, and confirmed that the expression of PMP genes are midgut specific, while the expression of other families of CBPs are more diverse in different tissues with substantial expression in cuticle forming tissues. Based on phylogenetic analysis of CBDs, CPAP1s form a separate cluster containing 16 different groups, including 5 new groups (groups K to O), and CPAP3s were clustered into 7 groups. In contrast, CBDs from the same PMPs appear to group together, indicating recent duplications of these domains. In CDAs and CHTs, the CBDs showed the same phylogenetic groups as those of the catalytic domains, with the exception of group IV CHTs.
Based on these results, we propose that CBPs are ancient proteins and that CBDs were duplicated and acquired by other proteins including enzymes of chitin metabolism. Two major groups with subfamilies evolved from ancestral CPAP1s and CPAP3s before the evolutionary radiation of insects. The CBDs in PMPs may have been initially derived from CPAP3s, followed by domain and gene multiplication events that occurred after divergence of the insect orders. For the chitin metabolism enzymes (CHTs and CDAs), the CBDs may have also been derived from CPAP3s by domain fusion before their diversification where CBDs evolved concomitantly with the catalytic domain. This study provides an extensive overview of CBPs and CBDs in proteins from M. sexta. With more insect genomes being sequenced, further analysis of these protein families from different species will provide additional data for understanding the evolutionary relations of the CBPs and their functional roles in ecologically, physiologically and developmentally diverse species of insects.
Supplementary Material
Acknowledgments
This project was supported in part by the Cornell University Agricultural Experiment Station federal formula funds received from the USDA Cooperative State Research, Education, and Extension Service to PW, and by the National Science Foundation through grant IOS-1022227 to SM. This is contribution #15-210-J from the Kansas Agricultural Experiment Station. The computing for this project was performed at the OSU High Performance Computing Center (OSUHPCC) at Oklahoma State University (OSU) which was supported in part through instrumentation funded by the National Science Foundation through grant OCI-1126330. Genome and RNA-seq data were provided by the Manduca Genome Project which was funded by a Defense Advanced Research Projects Agency grant to GB and a National Institutes of Health (GM041247) grant to MK.
Abbreviations:
- CDA
chitin deacetylase
- CBD
chitin binding domain
- CBP
chitin-binding protein
- CHT
chitinase
- CPAP
cuticular proteins analogous to peritrophins
- PM
peritrophic matrix (or peritrophic membrane)
- PMP
peritrophic matrix proteins (or peritrophic membrane proteins)
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ibmb.2014.12.002.
References
- Agrawal S, Kelkenberg M, Begum K, Steinfeld L, Williams CE, Kramer KJ, Beeman RW, Park Y, Muthukrishnan S, Merzendorfer H, 2014. Two essential peritrophic matrix proteins mediate matrix barrier functions in the insect midgut. Insect Biochem. Mol. Biol. 49, 24–34. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S, 2010. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 67, 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane Y, Zhu QS, Matsumiya M, Muthukrishnan S, Kramer KJ, 2003. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem. Mol. Biol. 33, 631–648. [DOI] [PubMed] [Google Scholar]
- Barry MK, Triplett AA, Christensen AC, 1999. A peritrophin-like protein expressed in the embryonic tracheae of Drosophila melanogaster. Insect Biochem. Mol. Biol. 29, 319–327. [DOI] [PubMed] [Google Scholar]
- Bass C, Zimmer CT, Riveron JM, Wilding CS, Wondji CS, Kaussmann M, Field LM, Williamson MS, Nauen R, 2013. Gene amplification and microsatellite polymorphism underlie a recent insect host shift Proc. Natl. Acad. Sci. U. S. A. 110, 19460–19465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr M, Hoch M, 2005. Identification of the novel evolutionary conserved obstructor multigene family in invertebrates. Febs Lett 579, 6827–6833. [DOI] [PubMed] [Google Scholar]
- Cates RG, 1980. Feeding patterns of monophagous, oligophagous, and polyphagous insect herbivores - the effect of resource abundance and plant chemistry. Oecologia 46, 22–31. [DOI] [PubMed] [Google Scholar]
- Cohen E, 2010. Chitin biochemistry: synthesis, hydrolysis and inhibition. In: Casas J, Simpson SJ (Eds.), Advances in Insect Physiology: Insect Integument and Color, pp. 5–74. [Google Scholar]
- Combet G, Blanchet C, Geourjon C, Deleage G, 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25,147–150. [DOI] [PubMed] [Google Scholar]
- Cornman RS, Willis JH, 2008. Extensive gene amplification and concerted evolution within the CPR family of cuticular proteins in mosquitoes. Insect Biochem. Mol. Biol. 38, 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE, 2004. WebLogo: a sequence logo generator. Genome Res. 14,1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer NT, Tetreau G, Cao X, Jiang H, Wang P, Kanost MR Annotation and expression analysis of cuticular proteins from the tobacco homworm, Manduca sexta. Insect Biochem. Mol. Biol, (in revision). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Arakane Y, Specht CA, Richard G, Kramer KJ, Beeman RW, Muthukrishnan S, 2008. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 38, 440–451. [DOI] [PubMed] [Google Scholar]
- Elvin GM, Vuocolo T, Pearson RD, East IJ, Riding GA, Eisemann CH, Tellam RL, 1996. Characterization of a major peritrophic membrane protein, peritrophin-44, from the larvae of Lucilia cuprina – cDNA and deduced amino add sequences. J. Biol. Chem. 271, 8925–8935. [DOI] [PubMed] [Google Scholar]
- Howe EA, Sinha R, Schlauch D, Quackenbush J, 2011. RNA-Seq analysis in MeV. Bioinformatics 27, 3209–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iconomidou VA, Willis JH, Hamodrakas SJ, 2005. Unique features of the structural model of ‘hard’ cuticle proteins: implications for chitin-protein interactions and cross-linking in cuticle. Insect Biochem. Mol. Biol. 35, 553–560. [DOI] [PubMed] [Google Scholar]
- Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ, 2014. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile Hidden Markov Models. Insect Biochem. Mol. Biol. 52, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S, 2010. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phytogeny, expression and function. Insect Biochem. Mol. Biol. 40, 214–227. [DOI] [PubMed] [Google Scholar]
- Jasrapuria S, Specht CA, Kramer KJ, Beeman RW, Muthukrishnan S, 2012. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. Plos One 7, e49844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KJ, Corpuz L, Choi HK, Muthukrishnan S, 1993. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manducasexta. Insect Biochem. Mol. Biol. 23, 691–701. [DOI] [PubMed] [Google Scholar]
- Kramer KJ, Muthukrishnan S, 2005. Chitin metabolism in insects. In: Gilbert LI, Iatrou K, Gill SS (Eds.), Comprehensive Molecular Insect Science. Elseviers B.V, Amsterdam, New York, pp. 111–144. [Google Scholar]
- Kuraishi T, Hori A, Kurata S, 2013. Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4, 375–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira CG, 2005. Invasion of the continents: cyanobacterial crusts to tree-inhabiting arthropods. Trends Ecol. Evol. 20, 253–262. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell G, Pop M, Salzberg SL, 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN, 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig S, Batz T, Armbruster K, Krasnow MA, 2006. serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 16,186–194. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki GJ., et al. , 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzendorfer H, Zimoch L, 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 206,4393–4412. [DOI] [PubMed] [Google Scholar]
- Petkau G, Wingen C, Jussen LCA, Radtke T, Behr M, 2012. Obstructor-a is required for epithelial extracellular matrix dynamics, exoskeleton function, and tubulogenesis. J. Biol. Chem. 287, 21396–21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers JE, Riddiford LM. 1988. Structure and expression of a Manduca-sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 203, 411–423. [DOI] [PubMed] [Google Scholar]
- Rebers JE, Willis JH, 2001. A conserved domain in arthropod articular proteins binds chitin. Insect Biochem. Mol. Biol. 31, 1083–1093. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Genta FA, Sorgine MH, Logullo R, Mesquita RD, Paiva-Silva GO, Majerowicz D, Medeiros M, Koerich L, Terra WR, Ferreira C, Pimentel AC, Bisch PM, Leite DC, Diniz MM, et al. , 2014. An insight into the transcriptome of the digestive tract of the bloodsucking bug, Rhodnius prolixus. Plos Neglected Trap. Dis. 8, e2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Stum A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, et al. , 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–+. [DOI] [PubMed] [Google Scholar]
- Shi XZ, Chamankhah M, Visal-Shah S, Hemmingsen SM, Erlandson M, Braun L, Alting-Mees M, Khachatourians GG, O’Grady M, Hegedus DD, 2004. Modeling the structure of the type I peritrophic matrix: characterization of a Mamestra configurata intestinal mucin and a novel peritrophin containing 19 chitin binding domains. Insect Biochem. Mol. Biol. 34, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Simonato M, Battisti A, Kerdelhue C, Burban G, Lopez-Vaamonde C, Pivotto I, Salvato P, Negrisolo E, 2013. Host and phenology shifts in the evolution of the social moth genus Thaumetopoea. PLoS One 8, e57192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam RL, Vuocolo T, Eisemann C, Briscoe S, Riding G, Elvin G, Pearson R, 2003. Identification of an immuno-protective mucin-like protein, peritrophin- 55, from the peritrophic matrix of Lucitta cuprina larvae. Insect Biochem. Mol. Biol. 33, 239–252. [DOI] [PubMed] [Google Scholar]
- Tellam RL, Wijffels G, Willadsen P, 1999. Peritrophic matrix proteins. Insect Biochem. Mol. Biol. 29, 87–101. [DOI] [PubMed] [Google Scholar]
- Terra WR, 2001. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch. Insect Biochem. Physiol. 47, 47–61. [DOI] [PubMed] [Google Scholar]
- Terra WR, Ferreira G, 2005. Biochemistry of digestion. In: Gilbert LI, Iatrou K, Gill SS (Eds.), Comprehensive Molecular Insect Science. Elseviers B.V, Amsterdam, New York, pp. 171–224. [Google Scholar]
- Tetreau G, Cao X, Chen Y-R, Muthukrishnan S, Haobo J, Blissard GW, Kanost MR, Wang P, 2015. Overview of chitin metabolism enzymes in Manduca sexto: identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 62,114–126. [DOI] [PubMed] [Google Scholar]
- Tiklova K, Tsarouhas V, Samakovlis G, 2013. Control of airway tube diameter and integrity by secreted chitin-binding proteins in Drosophila. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Granados RR, 1997. Molecular cloning and sequencing of a novel invertebrate intestinal mucin cDNA. J. Biol. Chem. 272, 16663–16669. [DOI] [PubMed] [Google Scholar]
- Wang P, Granados RR, 2001. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch. Insect Biochem. Physiol. 47, 110–118. [DOI] [PubMed] [Google Scholar]
- Wang P, Li GX, Granados RR, 2004. Identification of two new peritrophic membrane proteins from larval Trichoplusia ni: structural characteristics and their functions in the protease rich insect gut Insect Biochem. Mol. Biol. 34, 215–227. [DOI] [PubMed] [Google Scholar]
- Willis JH, 2010. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 40, 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Iconomidou VA, Smith RF, Hamodrakas SJ, 2005. Cuticular proteins. In: Gilbert LI, Iatrou K, Gill SS (Eds.), Comprehensive Molecular Insect Science. Elseviers B.V., Amsterdam, New York, pp. 79–109. [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ, 2012. Cuticular proteins. In: Gilbert LI (Ed.), Insect Molecular Biology and Biochemistry. Academic Press, San Diego, pp. 134–166. [Google Scholar]
- Zhu Q, Arakane Y, Baneijee D, Beeman RW, Kramer KJ, Muthukrishnan S, 2008. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem. Mol. Biol. 38, 452–466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


