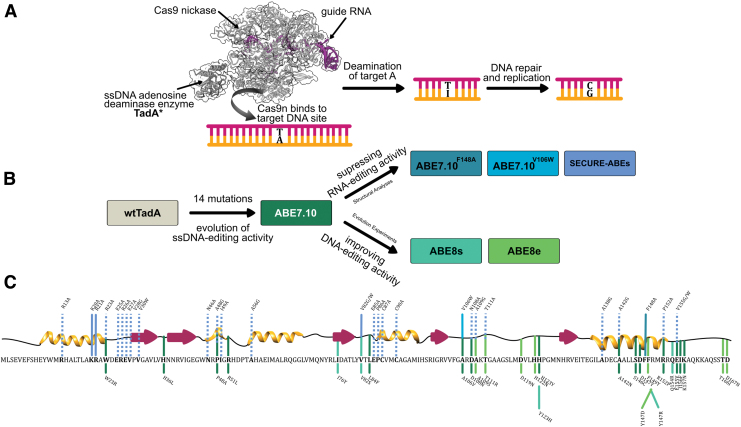
FIG. 1.
(A) Schematic representation of base editing by adenine base editors (ABEs; PDB ID: 6VPC).56 The binding of Cas9n to the target genomic locus unwinds the DNA double helix and exposes a small region of single-stranded DNA. TadA* hydrolytically deaminates the adenine (A) to form inosine (I), which is subsequently converted to guanine (G) by cellular DNA repair and replication machinery. (B) Engineering efforts in the field to generate and improve upon ABEs, starting from Escherichia coli wtTadA. (C) Primary and secondary structure of E. coli wtTadA with key mutations indicated. The line colors correspond to colors shown in (B), indicating the ABE version in which these mutations were identified. Solid lines are mutations that were incorporated into final ABEs, while dashed lines are mutations that were experimentally tested in previous work but not incorporated into final ABEs. Color images are available online.