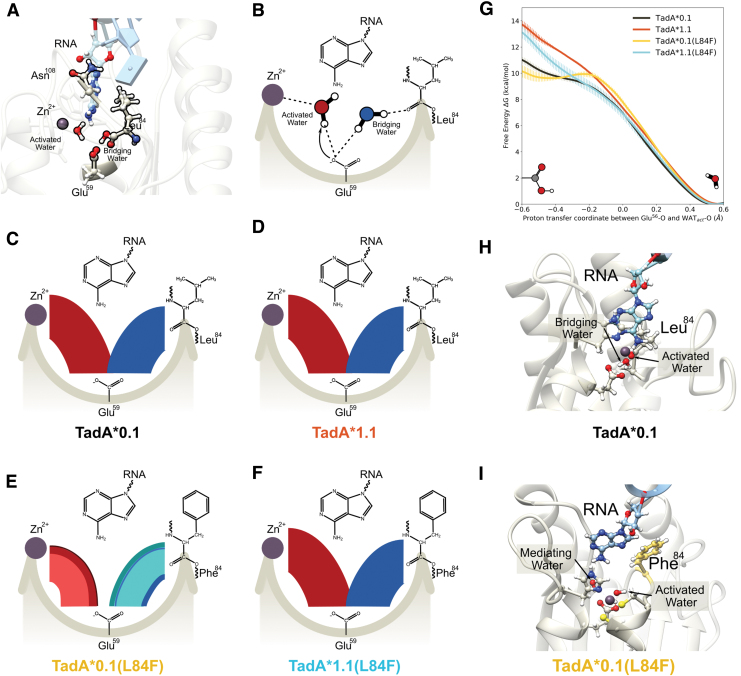
FIG. 6.
(A) Side view of the of the TadA*–RNA system highlighting the location of the catalytically relevant residues. The Zn+2 ion is coordinated by His57, Cys87, and Cys90 (not shown here for clarity) and a water molecule. This water molecule is activated by Glu59, which is also connected to another water molecule. This second water acts as a bridge between the Glu59 and the carbonyl backbone of residue 84. The target adenine is deep within the active site, and residue 108 is farther away from the active site waters. (B) Simplified flat lay representation to highlight the interactions of active site waters. Modified chord diagrams to demonstrate the persistence of the active site waters for (C) TadA*0.1– RNA, (D) TadA*1.1–RNII TadA*0.1(L84F)–RNA, and (F) TadA*1.1(L84F)–RNA. The red chords connecting Glu59 with Zn+2 depict the stability of the activated water molecule. Similarly, the blue chords connecting Glu59 with residue 84 depict the stability of the bridging water molecule. Different colors signify unique water molecules, with the thickness of individual chords being directly proportional to the total time these water molecules interact with the active site of TadA*–RNA during the simulation. (G) Reaction profile for the deprotonation of the activated water molecule the various TadA*–RNA systems. (H) Conformation of the TadA*0.1–RNA when the proton resides on Glu59. (I) Conformation of the TadA*0.1(L84F)–RNA when the proton resides on stability on the Glu59. The target A has moved back into the active site toward the Phe84 and is separated from the active site residues by an additional water molecule—the mediating water. Color images are available online.