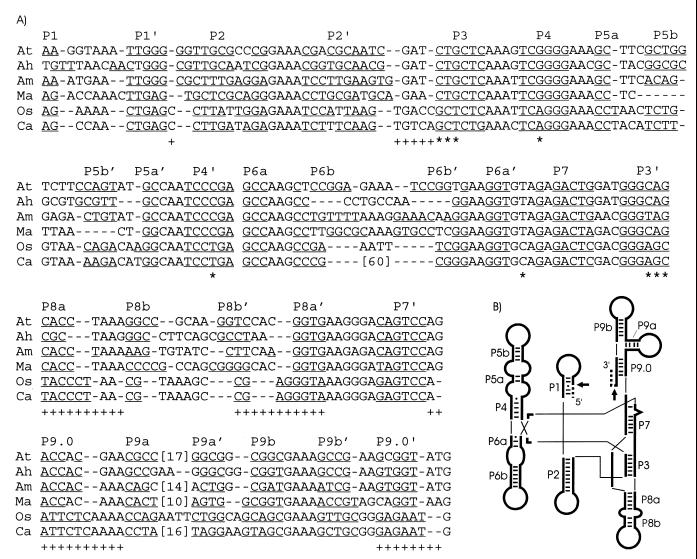
FIG. 1.
Intron sequence alignment and secondary structures. (A) DNA sequences of tRNACCUArg introns of the α-purple bacteria A. tumefaciens (At) (18), A. halopraeferens (Ah), and A. marginale (Am) are aligned with tRNAUAALeu introns from three distantly related cyanobacteria, M. aeruginosa (Ma) (19), Oscillatoria PCC 6304 (Os) (17), and Calothrix PCC 7101 (Ca) (17). Phylogenetically conserved base-paired regions (P1 to P9) (15) are indicated, with putative base-pairings underlined. Proposed base-pairing patterns of the Microcystis intron have been altered from the original report (19) to better conform to the conserved secondary structure of group I introns (15). Numbers in brackets indicate omitted sequence, and dashes represent gaps introduced to improve the alignment. Primary sequence (*) or secondary structural (+) similarities pointing to a closer relationship of the Microcystis tRNAUAALeu introns to the tRNACCUArg introns, rather than to the other known cyanobacterial tRNAUAALeu introns, are shown below the alignment. Note that these emphasized primary and secondary structural motifs are conserved among the cyanobacterial tRNAUAALeu introns (17), excluding the Microcystis introns. (B) Schematic representation of the consensus secondary structure of eubacterial tRNA group I introns, drawn according to Cech et al. (5). Phylogenetically conserved stems (P1 to P9), Watson-Crick base pairs (bars), and G-U pairs (dots) are shown according to Michel and Westhoff (15). Exons (dashed lines), the intron (thick lines), and splice sites (arrows) are indicated. Thin lines are used to join helical domains.