Figure 2.
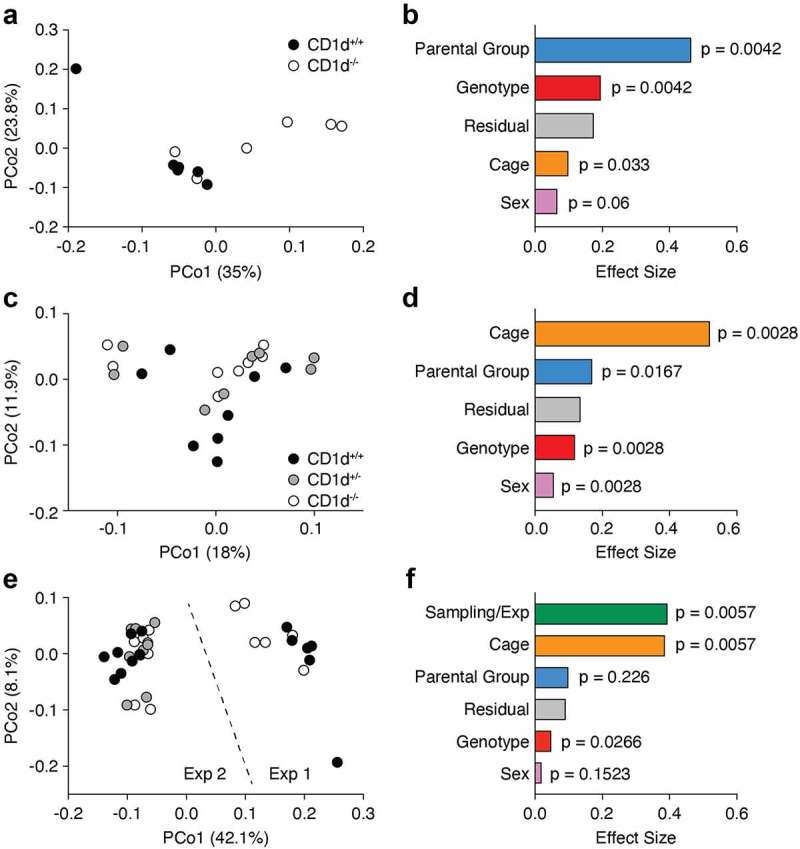
CD1d-deficiency and loss of iNKT cells does not alter microbiota composition. (a, b) 16S bacterial rRNA sequencing was used to define the microbiota profiles of CD1d+/+ and CD1d−/− littermate mice (n = 6 mice per group). These groups were separated by principal coordinates PCo1 and PCo2, collectively explaining 58.8% of the total similarity between samples, based on Bray-Curtis distances (a). Permutational multivariate analysis of the variance using Adonis (b). Data shows R2 (effect size) and adjusted p values for genotype, sex, caging and parental group. (c, d) 16S bacterial rRNA sequencing was used to define the microbiota profiles of CD1d+/+, CD1d+/− and CD1d−/− littermate mice (n = 8 mice per group) obtained from 2 consecutive litters from heterozygous harem breeding (1 male + 3 females). These groups were separated by principal coordinates PCo1 and PCo2, collectively explaining 29.9% of the total similarity between samples, based on Bray-Curtis distances (c). Permutational multivariate analysis of the variance using Adonis (d). Data shows R2 (effect size) and adjusted p values for genotype, sex, caging and parental group. (e, f) Combined Bray-Curtis PCoA of the two experiments (e) and Adonis (f) shows R2 (effect size) and adjusted p values for genotype, sex, caging, parental group and sampling/experiment.
