Figure 1. Modeling intracellular signaling and disease in context of “cue-signal-response” paradigm.
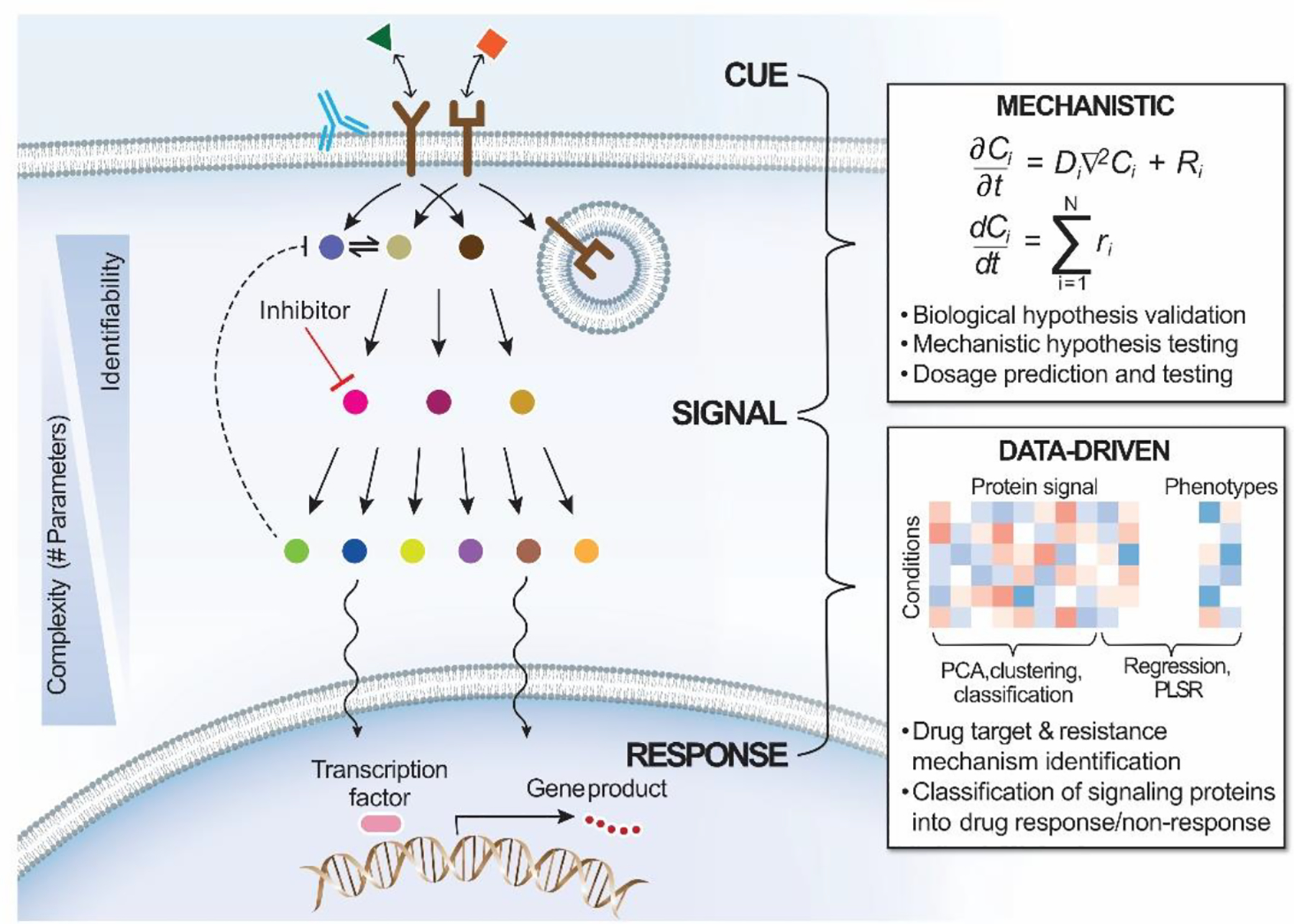
Intracellular signaling generally proceeds with a cue from the extracellular environment in form of ligands. Ligands bind to cognate receptors at the plasma membrane to activate receptors and downstream signaling pathways. Downstream effectors often undergo nucleocytoplasmic shuttling and regulate cell response via regulation of gene transcription. These processes are regulated by a variety of mechanisms, including receptor endocytic trafficking and feedbacks. In mechanistic models of signal transduction, biochemical reactions are typically represented by systems of differential equations, which include numerous parameters reflecting the rates of various binding and catalytic steps. These parameters determine model complexity and identifiability. Signaling dynamics and parameter relations deduced from mechanistic models help to form and validate biological hypotheses and can be applied to predict drug dosing regimens. Insight into signal-to-response relationships is more readily gained by data-driven models, which can reveal relevant structures in large signaling-relevant data sets and predict the importance of groups of signaling pathways in determining complex cell phenotypes. Common applications for data-driven models of signaling include identifying drug targets and resistance mechanisms.
