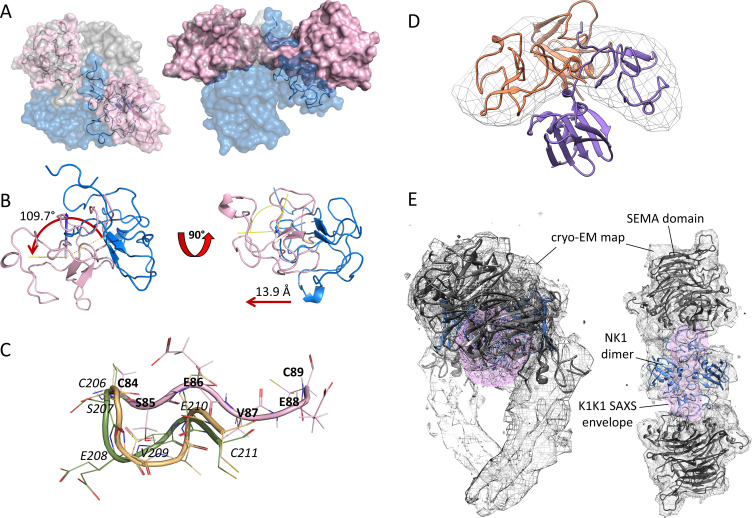
Figure S2. Structural alignment of K1K1 with NKI dimer.
(A) Semi-transparent surface presentation of K1K1 alignment based on one kringle domain with that of the kringle domain of one NK1 protomer. The alignment projects the second K1K1 kringle across and positions it close to but not in the same position as the second kringle domain in the NK1 dimer (blue). (B) Two detailed 90° views of the K1K1-NK1 kringle alignment showing the kringle of K1K1 in pink and the kringle of the NK1 protomer in magenta. The straight, stretched-out conformation of K1K1 misaligns the second kringle domain by a rotation of 109.7°, leading to a translation of 13.9 Å. (C) The linker based on the naturally occurring linker sequence between kringle 1 and kringle 2 in hepatocyte growth factor/scatter factor, SEVE, is straight in K1K1 (pink) and has a different conformation in two NK2 structures available (3HN4 in yellow and 3SP8 in green). Numbering is given in italic from the last cysteine of kringle 1 (C206 in NK2) until the first cysteine in kringle 2 (C211 in NK2) with equivalent K1K1 residues in bold. (D) Superimposition of K1K1 SAXS envelop with the two kringle domains of the protomers in the NK1 dimer. (E) Additional side and top views of the K1K1 SAXS envelope (pink) placed within the cryo-EM map of the NK1 dimer (blue)-SEMA domain (grey) complex (7mob.pdb).
Source data are available for this figure.