Fig. 1 |. Genome analysis of tandem repeats.
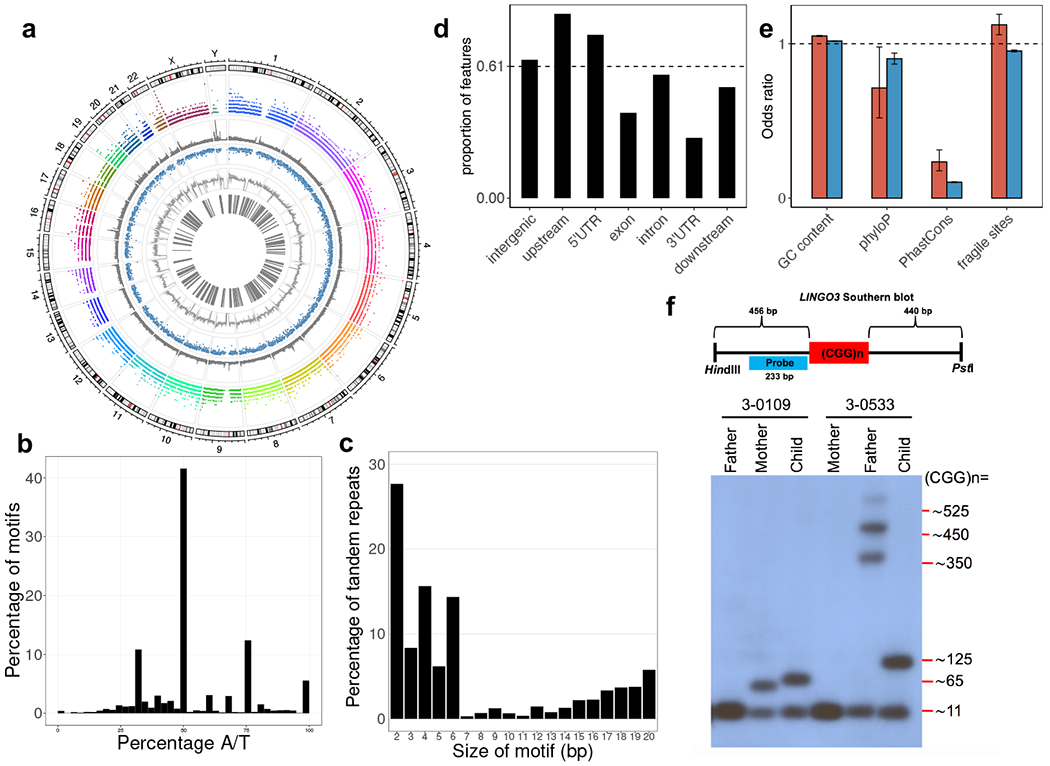
a, Circos plot showing the genomic distributions (1st layer) of 31,793 regions with tandem repeats (2nd layer), known simple sequence repeat regions (3rd layer), sequence conservation (4th layer), GC content (5th layer), and known fragile sites (6th layer). b, Nucleotide composition of the tandem repeats detected. c, Distribution of repeat unit (motif) sizes for the tandem repeats detected. d, Proportion of genic features overlapped by the tandem repeats detected. The proportion is derived from the size of tandem repeats over the total size of each genic feature. Dashed line indicates genome-average level. e, Correlation analysis between tandem repeats and different genomic features in a. By binning the genome into 1 kb windows, we tested the correlation/enrichment of different genomic features and the tandem repeats by regressing a genomic feature on the number of tandem repeats found per window. The odds ratios were derived from the logistic regression coefficients of the genomic features. Red bars represent tandem repeats detected (N=31,793 tandem repeat loci), while blue bars represent known simple sequence repeats (N=1,031,708 known short tandem repeats). Error bars indicate 95% confidence intervals. f, Validation of variable size in a tandem repeat detected. Schematic diagram (top) shows the design of a Southern blotting experiment in the targeted repeat in LINGO3, which overlaps with the location of fragile site FRA19B. Two families with different repeat sizes (3-0109 and 3-0533) are shown. In family 3-0533, the allele of size ~125 CGG repeats in the child appears to be a contraction of the father’s expanded allele, which displays multiple bands varying in repeat size (~350, ~450, and ~525 CGG repeats). Repeat length validation experiments for LINGO3 were consistently reproduced at least 3 to 5 times (see Supplementary Figure 8).
