Extended Data Figure 1 |. Study design.
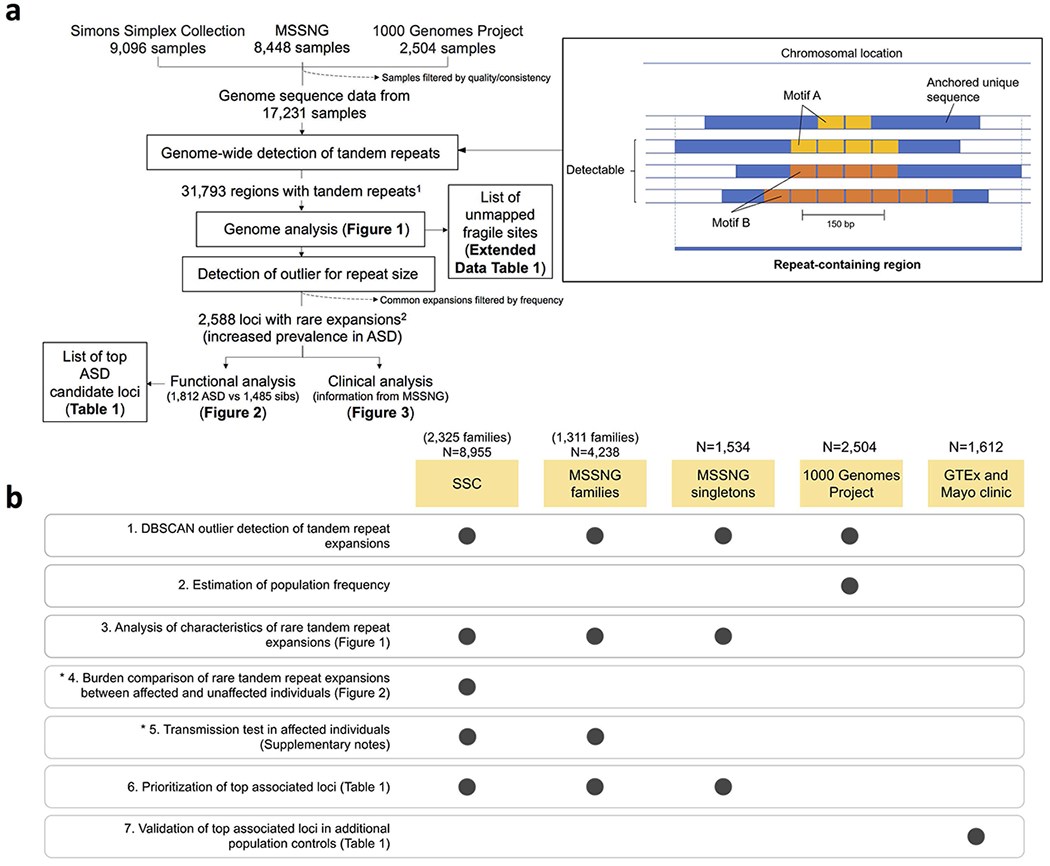
a, Schematic workflow of the tandem repeat detection and analyses. 1Tandem repeats here are defined as those with 2–20 bp repeat motifs that span at least 150 bp. 2Rare expansions here are defined as tandem repeat expansions that are outliers according to size and occur in <0.1% of population controls from the 1000 Genomes Project. Note that ExpansionHunter Denovo only approximates the size and location of a given tandem repeat; thus, we use the term “region” to refer to a genomic segment detected in this way, and reserve “location” or “locus” for sites that have been more precisely mapped. b, Genome sequencing cohorts used for each analysis performed in this study. Numbers above each cohort represent the number of samples remaining after curation (Supplementary Notes).
