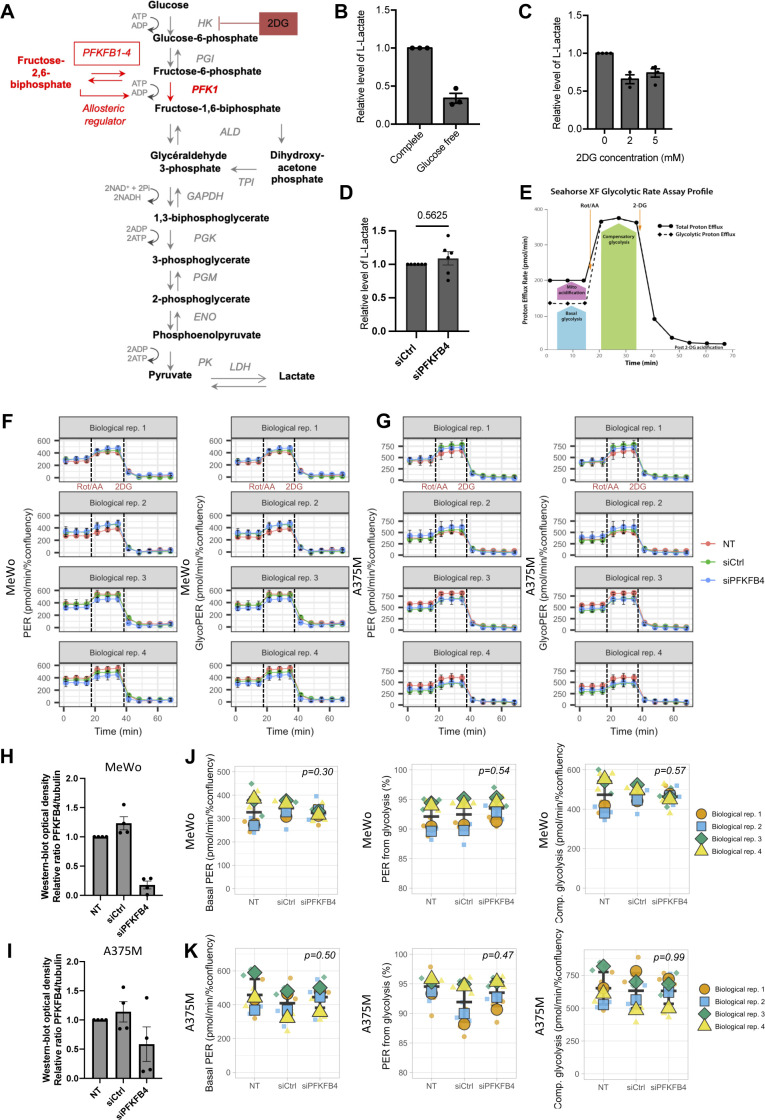
Figure S3. PFKFB4 controls metastatic melanoma cell migration in a glycolysis-independent manner.
(A) Summary of glycolysis. The rate-limiting step of glycolysis is the second irreversible reaction, catalyzed by phosphofructokinase-1 (PFK1). PFK1 activity depends on the availability of its allosteric regulator, fructose-2,6-bisphosphate. PFKFB4 catalyzes the synthesis or degradation of fructose-2,6-bisphosphate. The 2-deoxyglucose (2DG) is a glucose analog that cannot be metabolized. 2DG blocks glycolysis by competition with cellular glucose. At the end of the glycolysis, pyruvate is transformed into lactate, which is secreted in the extracellular medium. Lactate levels are a readout for the rate of glycolysis. (B, C, D) Relative extracellular L-lactate levels in MeWo cells cultured either in a complete medium or in glucose-free medium (B), treated with different concentrations of 2DG (C) or transfected either with siCtrl or siPFKFB4 (D). P-value was calculated using the Mann–Whitney test. (E) Seahorse XF glycolytic rate assay profile highlighting different glycolytic parameters measurement after sequential addition of rotenone/antimycin A mitochondrial respiration inhibitors and 2-DG glycolysis inhibitor. (F, G) Real-time proton efflux rate (PER) and glycoPER calculated from extracellular acidification rate and oxygen consumption rate measurements performed and analyzed by Seahorse in MeWo (F) and A375M (G) cells, in four different biological replicates non transfected (NT, red), or transfected with siCrl (green) or siPFKFB4 (blue). Bars represent mean ± SD of technical replicates. (H, I) Normalized PFKFB4/tubulin levels in MeWo (H) or A375M (I) determined by Western blot of the four biological replicates used for the Seahorse experiments. One point represents one biological replicate. (F, G, J, K) SuperPlots of mean values of the total basal PER, percentage of glycoPER, and compensatory glycolysis deduced from real-time Seahorse experiments presented in (F) and (G) for MeWo (J) and A375M (K) cells. Each small shape represents a technical replicate, and large shapes represent the mean value of each biological replicate. Bars represent mean ± SD. P-value was calculated using t test.