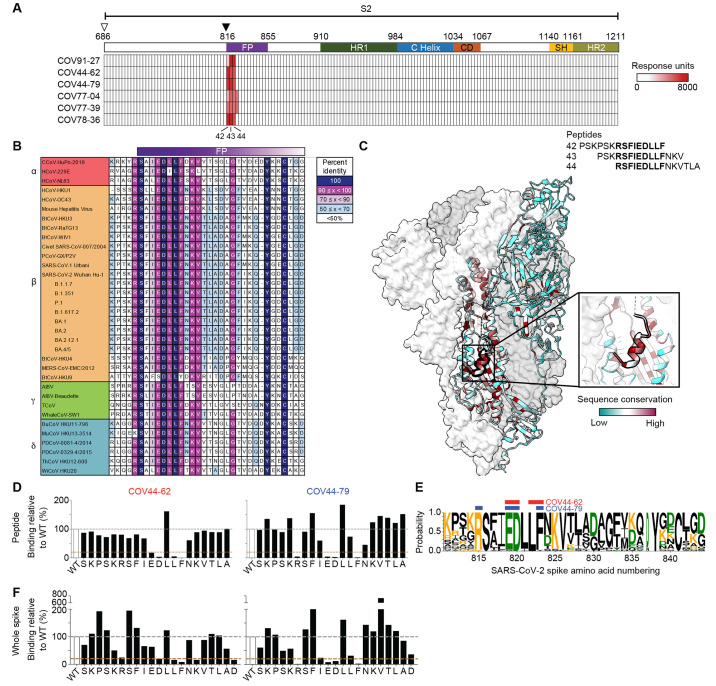
Fig. 3. Broadly neutralizing antibodies target the conserved fusion peptide.
(A) Heat map of SARS-CoV-2 S2 peptide array. Binding responses were assessed by SPR using a 15-mer peptide array with 12 aa overlay covering the entire S2 subunit. Each column within the map represents a single peptide. The white triangle shows the S1/S2 cleavage site and the black triangle indicates the S2' cleavage site. FP, fusion peptide; HR1, heptad repeat 1; C helix, central helix; CD, connector domain; SH, stem helix; HR2, heptad repeat 2. (B) Sequence alignment of the fusion peptide from 34 viral isolates representing a diverse group of coronaviruses across four genera. Performed using MAFFT v7.450 using a BLOSUM62 scoring matrix and the L-INS-I algorithm. (C) Sequence conservation of pre-fusion SARS-CoV-2 spike protein (PDB 6VSB) with the fusion peptide (aa 816-843) highlighted in a black outline. Inset shows a magnified view of this region. (D) Alanine scan evaluating the binding of COV44-62 and COV44-79 to the SARS-CoV2 fusion peptide. Responses were normalized to the wild-type sequence. A cut-off of 20% (brown hashed line) was used to identify residues that were critical for binding. (E) Sequence logo plot of diversity within the fusion peptide region of coronaviruses from 34 isolates, built using WebLogo 3. Height is proportional to the probability of an amino acid at a given position and amino-acid residues are colored by charge. Narrow stacks (amino acids) indicate deletions or gaps in the sequences. Numbering is based on the SARS-CoV-2 Wuhan-Hu-1 sequence. The key residues in the epitope footprints of mAbs COV44-62 (red) and COV44-79 (blue), based on peptide alanine scanning, are highlighted above the logo plot. (F) Amino acids critical for the binding of COV44-62 and COV44-79 identified by shotgun alanine mutagenesis of S2 residues on whole spike protein. Only fusion peptide residues are shown here. Key residues were identified based on a <20% signal relative to wild-type spike (brown hashed line), with no corresponding loss of signal for a control mAb, which targets the spike protein but does not bind to this site (see fig. S3C).