Abstract
Objective
To identify and characterize genetic loci associated with the risk of developing ANCA-associated vasculitides (AAV).
Methods
Genetic association analyses were performed after Illumina sequencing of 1853 genes and subsequent replication with genotyping of selected single nucleotide polymorphisms in a total cohort of 1110 Scandinavian cases with granulomatosis with polyangiitis or microscopic polyangiitis, and 1589 controls. A novel AAV-associated single nucleotide polymorphism was analysed for allele-specific effects on gene expression using luciferase reporter assay.
Results
PR3-ANCA+ AAV was significantly associated with two independent loci in the HLA-DPB1/HLA-DPA1 region [rs1042335, P = 6.3 × 10−61, odds ratio (OR) 0.10; rs9277341, P = 1.5 × 10−44, OR 0.22] and with rs28929474 in the SERPINA1 gene (P = 2.7 × 10−10, OR 2.9). MPO-ANCA+ AAV was significantly associated with the HLA-DQB1/HLA-DQA2 locus (rs9274619, P = 5.4 × 10−25, OR 3.7) and with a rare variant in the BACH2 gene (rs78275221, P = 7.9 × 10−7, OR 3.0), the latter a novel susceptibility locus for MPO-ANCA+ granulomatosis with polyangiitis/microscopic polyangiitis. The rs78275221-A risk allele reduced luciferase gene expression in endothelial cells, specifically, as compared with the non-risk allele.
Conclusion
We identified a novel susceptibility locus for MPO-ANCA+ AAV and propose that the associated variant is of mechanistic importance, exerting a regulatory function on gene expression in specific cell types.
Keywords: ANCA-associated vasculitis, PR3-ANCA, MPO-ANCA, genetic analysis, BACH2, regulatory variant
Rheumatology key messages.
A single nucleotide polymorphism at the BACH2 locus was identified as significantly associated with MPO-ANCA+ AAV.
The rare genetic variant exerted a cell type-specific regulatory function.
Introduction
Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitides (AAV) are rare, potentially life-threatening diseases, characterized by necrotizing inflammation of small- and medium-sized blood vessels. AAV have traditionally been categorized into the clinical entities granulomatosis with polyangiitis (GPA), microscopic polyangiitis (MPA) and eosinophilic GPA (EGPA). EGPA is clearly distinct, characterized by, e.g. hypereosinophilia. GPA and MPA, on the other hand, share some clinical and pathological features and differ in others, such as increased involvement of upper airways and presence of granulomas in GPA and a higher incidence of renal involvement in MPA. Additionally, GPA is mainly associated with auto-antibodies (ANCAs) against proteinase 3 (PR3), whereas MPA to a larger degree is associated with myeloperoxidase (MPO)-ANCA [1, 2].
Emerging evidence from studies of genetic predisposition and environmental triggers of disease propose that GPA and MPA are distinct diseases with different aetiological mechanisms [3–5]. Two genome-wide association studies (GWAS) have identified unique genetic susceptibility loci for GPA (HLA-DPB1/DPA1, PRTN3, SERPINA1) and MPA (HLA-DQB1/DQA1) [3, 4]. Notably, these studies revealed a stronger association between the genetic loci and the presence of PR3- and MPO-ANCA, respectively, than with the clinical diagnoses, hence challenging the current categorization of AAV.
Commonly used single nucleotide polymorphism (SNP) array-based GWAS are well suited for identification of common genetic variants associated with disease, but lack efficiency in revealing information about rare variation, determining causative variants and in prioritizing non-protein coding regions [6]. These limitations may be overcome by using DNA sequencing, whereby common as well as rare variants are detected, increasing the chances of identifying the actual causative alleles.
In the present study, we sought to increase the current understanding of the pathogenesis of AAV through identification of common and rare genetic variation associated with PR3- and MPO-ANCA+ AAV. Using a resequencing approach targeting almost 1900 immune-related genes, we were able to cover both coding and selected non-coding regions of the genome and to identify disease-associated genetic variants in a homogeneous cohort of Scandinavian AAV patients.
Methods
For full information on methods, see the online Supplementary Methods, available at Rheumatology online.
Subjects
All cases included in the analyses of the present study were clinically diagnosed with GPA or MPA and met the corresponding classification criteria according to the European Medicines Agency algorithm [7]. Patients were recruited via six university hospitals in Sweden and Norway for the discovery cohort (n = 679) and via eight hospitals in Sweden and Denmark for the replication cohort (n = 536), and were included in the study after informed and written consent. Healthy controls were recruited from blood donors or population controls from Sweden and Norway (n total = 1706). The study complies with the declaration of Helsinki. The locally appointed ethics committee approved the research protocol.
Genetic variant discovery
Sequence capture and DNA sequencing were performed with samples from patients of the discovery cohort and from controls of the discovery and replication cohort. The sequence capture of the exons of 1853 genes with relevance for immune-mediated disease [8] and evolutionarily conserved regions within 100 000 bps of the genes, was designed and applied using SeqCap EZ Choice XL (Roche NimbleGen, Pleasanton, CA, USA) and the DNA products were sequenced on Illumina HiSeq 2500, using a protocol described previously [8]. Quality control (QC) of sequence data with filtering of samples and variants is summarized in supplementary Fig. S1, available at Rheumatology online (for details, see online Supplementary Methods). After QC, a total of 602 out of the originally recruited 679 patient samples (PR3-ANCA+: n = 425; MPO-ANCA+: n = 175; ANCA negative: n = 15; supplementary Table S1, available at Rheumatology online) remained for analysis as a discovery cohort. For the healthy controls, 1597 out of the 1706 originally recruited individuals remained after filtering of sequence data; 999 of these were selected as a control population for the patient discovery cohort, with the remaining 597 individuals saved for the replication analysis (n = 590 after additional QC; supplementary Table S1, available at Rheumatology online). In the whole discovery data set of 1601 individuals, 359 013 SNPs [115 291 with minor allele frequency (MAF) > 0.01] passed QC (supplementary Table S2, available at Rheumatology online).
For the replication study, iPLEX MassARRAY (Agena Bioscience, San Diego, CA, USA) was used for genotyping of 37 selected SNPs in 536 patient samples. After QC of variants and samples, 31 SNPs remained for analysis in 508 cases (PR3-ANCA+: n = 401; MPO-ANCA+: n = 101; ANCA negative: n = 8). The corresponding SNPs were analysed in targeted sequence data of the healthy controls (n = 590 after QC). There was a >99% genotype concordance rate between sequenced and MassARRAY genotyped candidate SNPs in 41 individuals that were analysed using both methods (supplementary Fig. S2, available at Rheumatology online).
Statistical analysis
Single-variant association analysis was performed using logistic regression analyses (Plink v.1.9) of PR3-ANCA+ and MPO-ANCA+ patients, separately, against controls, in both the discovery and replication cohorts. Meta-analyses of the discovery and replication data sets, with PR3-ANCA+ and MPO-ANCA+ patients separately, were performed using GWAMA v2.2.2 [9]. After estimation of the number of independent tests [SNPs were considered dependent if linkage disequilibrium (LD) r2 > 0.8], the P-value threshold for significance for the analysis of the discovery cohort and the meta-analysis was set to P < 9.1 × 10−7.
Gene-based aggregate tests were performed on the discovery data set using SKAT-O in R v.3.4.1 (package Skat v.1.3.2.1), with a Bonferroni-adjusted P-value threshold of <2.8 × 10−5 (P < 0.05 adjusted for 1793 tests).
Allele-specific expression analysis
The allele-specific effects of rs78275221 on gene expression were analysed using luciferase reporter assays, by insertion of a 161-bp fragment centered on rs78275221, with each of the two alleles (G/A) into separate pGL4.26 vectors (Promega, Madison, WI, USA). Vectors were transfected into Jurkat, Daudi and human dermal microvasculature endothelial (HMVEC) cells, respectively, and the effects of the allele-specific fragments on luciferase activity (Jurkat and Daudi cells) and luciferase transcript levels (HMVEC), were assayed. Allele-specific differences in gene expression were analysed using unpaired Student’s t-test. mRNA expression of BACH2 in Jurkat, Daudi and HMVEC cells was analysed using reverse transcriptase PCR.
Results
Single-variant association analysis identifies known and novel AAV-associated loci
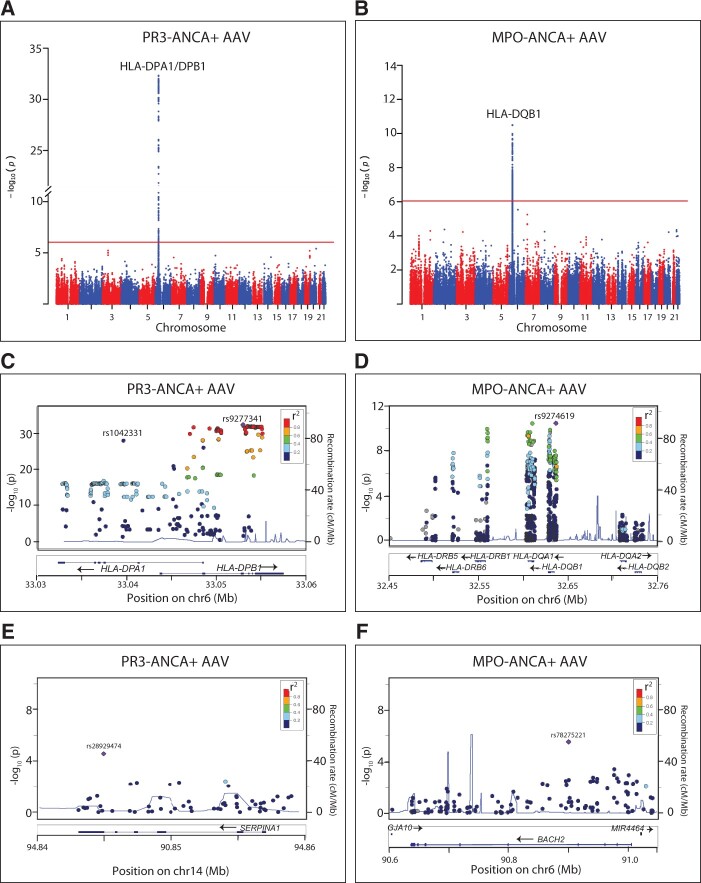
Based on previous findings [3, 4], we decided to consider AAV as two separate diseases based on ANCA status: PR3-ANCA+ and MPO-ANCA+ AAV. Single-variant association analysis of SNPs with MAF >0.01 in the discovery dataset identified a total of 335 SNPs and 168 SNPs as significantly associated with PR3-ANCA+ AAV and MPO-ANCA+ AAV, respectively, all located in the HLA region (Fig. 1A and B; supplementary Tables S3 and S4, available at Rheumatology online). In PR3-ANCA+ AAV, conditional analysis revealed two independent signals of association in the HLA region, with the lead SNPs rs1042331 in HLA-DPB1 [P = 4.6 × 10−33, odds ratio (OR) 0.088 (95% CI 0.059, 0.13)] and rs9277341 in HLA-DPA1 [P = 8.9 × 10−29, OR 0.19 (0.14–0.25); Fig. 1C, Table 1]. In the MPO-ANCA+ cohort, one independent signal of association was identified in the HLA region, with lead SNP rs9274619 located in HLA-DQB1 [P = 3.2 × 10−11, OR 3.3 (2.3–4.7); Fig. 1D, Table 1]. In addition to the significantly associated loci, outside the HLA region 18 SNPs in 10 loci in the PR3-ANCA+ cohort and 14 SNPs in 9 loci in the MPO-ANCA+ cohort achieved P-values with a suggestive association to disease (P < 1.0 × 10−4; supplementary Tables S3–S5, available at Rheumatology online). SNPs at the PRTN3 locus (previously associated with PR3-ANCA+ AAV [3, 4]) were filtered out during QC due to unequal missingness between cases and controls (P < 3.4 × 10−11), hence the gene was not among the candidate associated loci.
Fig. 1.
Manhattan plots of the discovery analyses of PR3- and MPO-ANCA+ AAV
(A, B) The –log10P-values for all SNPs of the discovery analyses plotted against their chromosomal locations, with P-value threshold (9.1 × 10−7) indicated by a red line. (C–F) Locus zoom plots depict close-ups of the HLA-DP region, HLA-DQ region, SERPINA1 region and BACH2 region, respectively. (A, C, E) PR3-ANCA+ AAV; (B, D, F) MPO-ANCA+ AAV. Lead SNPs are labelled and remaining SNPs are coloured according to LD with lead SNP (r2). Blue lines = recombination rates; gene locations are assigned below plots. AAV: ANCA-associated vasculitides; LD: linkage disequilibrium; SNP: single nucleotide polymorphism.
Table 1.
Single-variant association analysis of discovery, replication and combined cohorts, with patients stratified according to presence of ANCA
| Discovery analysis (n PR3-ANCA+ = 425, MPO-ANCA+ = 175, controls = 999) |
Replication analysis (n PR3-ANCA+ = 401, MPO-ANCA+ = 101, controls = 590) |
Meta-analysis (n PR3-ANCA+ = 826, MPO-ANCA+ = 276, controls = 1589) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Position | SNP | Gene(s) | Minor allele | MAF cases | MAF controls | P | OR | 95% CI | MAF cases | MAF controls | P | OR | 95% CI | P | OR | 95% CI | |
| PR3-ANCA+ AAV | ||||||||||||||||||
| 6 | 33052950 | rs1042331 | HLA-DPB1 | C | 0.036 | 0.27 | 4.7 × 10–33 | 0.088 | 0.059, 0.13 | – | – | – | – | – | – | – | – | |
| 6 | 33052958 | rs1042335a | HLA-DPB1 | T | 0.038 | 0.27 | 5.4 × 10–33 | 0.094 | 0.064, 0.14 | 0.047 | 0.28 | 9.8 × 10–30 | 0.11 | 0.075, 0.16 | 6.3 × 10–61 | 0.10 | 0.077, 0.13 | |
| 6 | 33039625 | rs9277341 | HLA-DPA1 | C | 0.072 | 0.29 | 1.2 × 10–28 | 0.19 | 0.14, 0.25 | 0.080 | 0.270 | 4.2 × 10–18 | 0.26 | 0.19, 0.35 | 1.5 × 10–44 | 0.22 | 0.18, 0.27 | |
| 14 | 94844947 | rs28929474 | SERPINA1 | T | 0.055 | 0.023 | 2.6 × 10–5 | 2.6 | 1.7, 4.2 | 0.075 | 0.025 | 2.0 × 10–5 | 3.2 | 2.0, 5.3 | 2.7 × 10–10 | 2.9 | 2.1, 4.0 | |
| MPOANCA+ AAV | ||||||||||||||||||
| 6 | 32635954 | rs9274619 | HLA-DQB1, HLA-DQA2 | A | 0.34 | 0.14 | 3.2 × 10–11 | 3.3 | 2.3, 4.7 | 0.40 | 0.130 | 1.7 × 10–17 | 4.0 | 2.9, 5.7 | 5.4 × 10–25 | 3.7 | 2.9, 4.7 | |
| 6 | 90900544 | rs78275221 | BACH2 | A | 0.074 | 0.021 | 2.9 × 10–6 | 3.5 | 2.1, 5.9 | 0.050 | 0.0270 | 0.053 | 2.1 | 0.99, 4.5 | 7.9 × 10–7 | 3.0 | 1.9, 4.6 | |
Proxy for lead SNP rs1042331 that failed genotyping in the replication analysis.
SNP: single nucleotide polymorphism; OR: odds ratio; MAF: minor allele frequency; AAV: ANCA-associated vaculitis. PR3-ANCA+/MPO-ANCA+: AAV patients positive for indicated ANCA.
The top HLA SNPs identified in the discovery data analyses, a selected set of additional SNPs within the HLA region, and one to two SNPs representing each non-HLA locus with suggestive association with disease (see Supplementary Methods, available at Rheumatology online for details) were taken forward for genotype analysis in an independent cohort of subjects (Fig. 1E and F; supplementary Table S5, available at Rheumatology online). The PR3-ANCA-associated lead SNP rs1042331 failed genotyping but was represented by SNP rs1042335, in complete LD (r2 = 1.0, D′ = 1.0). All HLA SNPs, as well as four additional loci (SERPINA1, OGFR, RAPGEF5, RCN3), achieved a P-value of ≤0.05 in the analysis of PR3-ANCA+ AAV. Three SNPs at the HLA-DR/HLA-DQ locus and one SNP at the BACH2 locus achieved P ≤ 0.05 in the MPO-ANCA+ cohort (Table 1; supplementary Table S5, available at Rheumatology online).
Next, SNPs with P ≤ 0.05 of the replication data set were combined with the discovery data set in a meta-analysis. The strongest associations were identified between the independent signals of the HLA-DPB1 and HLA-DPA1 loci and PR3-ANCA+ AAV [rs1042335, P = 6.3 × 10−61, OR 0.10 (0.077–0.13); and rs9277341, P = 1.5 × 10−44, OR 0.22 (0.18–0.27), respectively]. In addition, there was a significant association between PR3-ANCA+ AAV and the SERPINA1 locus [rs28929474 (Z allele), P = 2.7 × 10−10, OR 2.9 (2.1–4.0); Table 1, Fig. 1E;supplementary Table S5, available at Rheumatology online]. The strongest signal of association with MPO-ANCA+ AAV was found at HLA-DQB1 [rs9274619, P = 5.4 × 10−25, OR 3.7 (2.9–4.7)]. Additionally, the SNP located at the BACH2 locus (rs78275221) was significantly association with disease [P = 7.9 × 10−7, OR 3.0 (1.9–4.6); Table 1, Fig. 1F;supplementary Table S5 and Fig. S3, available at Rheumatology online].
Finally, we performed a meta-analysis of the discovery and replication cohorts with the patients subcategorized according to their clinical diagnoses GPA and MPA, as well as all patients jointly. As expected, the PR3-ANCA-associated HLA-DPB1/DPA1 and SERPINA1 loci were also associated with GPA but with weaker signals [rs1042335, P = 2.9 × 10−60, OR 0.14 (0.11–0.17); rs9277341, P = 2.8 × 10−39, OR 0.28 (0.23–0.34); rs28929474, P = 5.8 × 10−10, OR 2.8 (2.0–3.8)]. For MPA, there was a significant association with the HLA-DQB1 locus but not with BACH2 [rs9274619, P = 1.9 × 10−17, OR 3.2 (2.4–4.2); rs78275221, P = 1.4 × 10−4, OR 2.5 (1.6–4.1); supplementary Tables S6–S8, available at Rheumatology online]. In the discovery analysis of the total AAV cohort, there were two independent significantly associated loci at HLA-DPB1 [rs9277469, P = 2.8 × 10−28, OR 0.28 (0.22–0.34)] and HLA-DQB1 [rs1770, P = 1.2 × 10−7, OR 1.6 (1.4–2.0)] (supplementary Table S6 and S9, available at Rheumatology online), but the signals were weaker than for the ANCA-specific cohorts. SERPINA1 (rs28929474) was the only non-HLA locus with a significant association with AAV in the meta-analysis [P = 5.0 × 10–8, OR 2.4 (1.7–3.2); supplementary Table S6, available at Rheumatology online].
Taken together, our results confirmed previous genetic associations with PR3- and MPO-ANCA+ AAV and identified a novel locus associated with MPO-ANCA+ AAV.
Rare variant aggregate test confirms association to the HLA region
In order to investigate a combined contribution of rare genetic variants to PR3-ANCA+ and MPO-ANCA+ AAV, the discovery data set was analysed using the gene-based aggregate test SKAT-O. To exclude the effects of common variants, only SNPs with a MAF <0.05 were included in the analysis. For PR3-ANCA+ AAV, significant associations were identified for four loci in the HLA class II region, whereas no regions were significantly associated with MPO-ANCA+ AAV (supplementary Table S10, available at Rheumatology online).
Functional evaluation of disease-associated genetic variants
At the novel MPO-ANCA+ AAV susceptibility locus BACH2, the significantly associated variant rs78275221 (G/A) is intronic and rare [MAF: Europeans 0.021 [GnomAD: CEU (Northern/Western European ancestry)] [10], Swedes 0.028 (Swefreq) [11]]. It is in strong LD with SNPs previously associated with several autoimmune disorders [2, 8, 12–14] and with an SNP with a previous suggestive association with MPO-ANCA+ AAV [4] (supplementary Table S11 and Fig. S4, both available at Rheumatology online). The position of rs78275221 is not evolutionarily conserved (PhyloP score –0.57), but is located in a region of open chromatin in B cells and T helper 1 cells, also characterized by enhancer-associated histone modifications (H3K27Ac, H3K4me1) in B cells, CD14+ monocytes and human umbilical vein endothelial cells [Encyclopedia of DNA elements (ENCODE) supplementary Fig. S4, available at Rheumatology online]. rs78275221 is located in a predicted binding motif of Ras-responsive element binding protein 1 (RREB1; JASPAR transcription factor binding site database [15]), where the A allele contributes to a higher affinity to RREB1 (P = 0.00084) compared with the G allele (P = 0.027; Transcription factor affinity prediction 0.011 vs 0.002 [16]; supplementary Fig. S4, available at Rheumatology online). There are three protein-coding genes within the topologically associated domain (TAD; BACH2, MAP3K7, GJA10) and suggestive data of physical interactions between the rs78275221 locus and the BACH2 promoter region, as well as to other regulatory regions within the TAD (supplementary Fig. S4, available at Rheumatology online) [17].
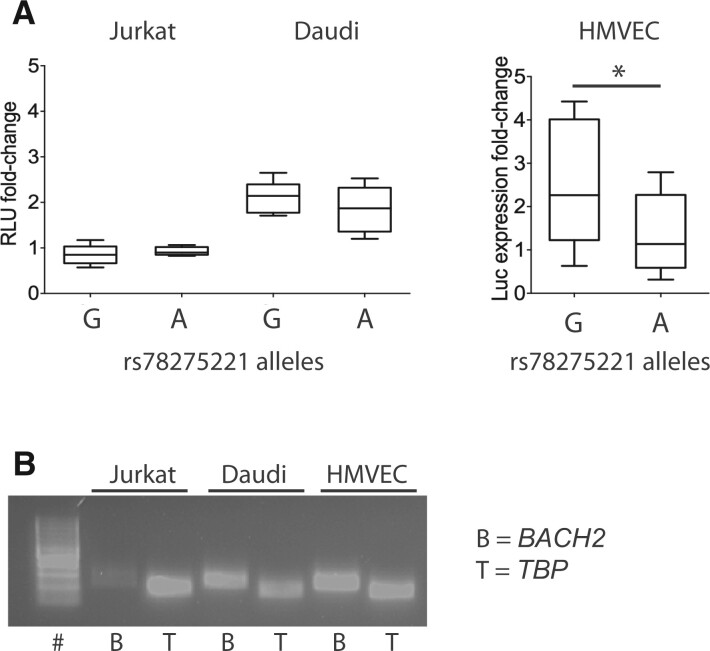
To investigate whether rs78275221 may exert a regulatory function, the effects of the two alleles on gene expression were analysed in T and B cells (Jurkat and Daudi cell lines, respectively) as well as in primary HMVEC using luciferase reporter assay. The rs78275221 locus did not affect luciferase expression in Jurkat cells (P = 0.15 and P = 0.19 for the G and A allele, respectively) but increased the expression two-fold in Daudi cells (P = 0.00010 and P = 0.00070 for the G and A allele, respectively), compared with vectors without insert (Fig. 2A). In HMVEC, insertion of rs78275221-G increased luciferase expression two-fold (P = 0.0040), whereas inserts with the disease risk allele rs78275221-A did not affect expression (P = 0.20) compared with vectors without insert, thereby revealing a significant difference between the two alleles in HMVEC (P = 0.02; Fig. 2A).
Fig. 2.
Allele-specific effects on gene expression of rs78275221 in BACH2-expressing cell types
(A) The allelic effects of rs78275221 on reporter expression are presented as RLU fold-change for Jurkat and Daudi cells and luciferase transcript expression fold-change for primary HMVEC, relative to the respective levels of the reporter vector without insert. Inserts with either of the two rs78275221 alleles were compared. Box plots illustrate median, first and third quartiles, minimum and maximum values. *P < 0.05 (unpaired t-test). (B) BACH2 mRNA expression analysis in Jurkat and Daudi cell lines and HMVEC, as analysed by reverse-transcriptase PCR and gel electrophoresis. # = 100-bp ladder. TBP was used as internal control. HMVEC: human dermal microvasculature endothelial; RLU: relative light units; TBP: TATA-box binding protein.
The cell type-specific expression of BACH2 is not fully known, but reverse transcriptase PCR analysis confirmed expression in Daudi cells and HMVEC but indicated low or minimal expression in Jurkat cells (Fig. 2B).
The lead SNPs of the HLA loci identified for PR3-ANCA+ and MPO-ANCA+ AAV in this study are in strong LD with lead HLA variants of previous GWAS [rs1042331, r2 = 0.72–1.0, D′ = 0.91–1.0; rs9277341, r2 = 1.0, D′ = 1.0 (identical SNP); rs9274619, r2 = 0.30–1.0, D′ = 1.0] [3, 4, 18]. HaploReg [19] predictions of potential functionality of associated HLA SNPs suggested numerous functional coding and non-coding variants at each locus, each with the potential to affect one or several transcripts (supplementary Tables S12 and S13, available at Rheumatology online). Intersection of the location of all SNPs in complete LD (r2 = 1, D′ = 1) with the lead HLA SNPs with ENCODE epigenetic data, implied coding or regulatory functions for a substantial proportion of the SNPs (supplementary Fig. S5, available at Rheumatology online).
Candidate genes and traditional cardiovascular disease
The presence of at least two well-known stroke-associated loci (SERPINA1, HDAC9) [20–24] in our list of loci with association or suggestive association with AAV in the discovery data set prompted us to investigate plausible associations between these candidate AAV genes and traditional cardiovascular disease (CVD). As risk variants may exert a regulatory function on both proximal and distal genes, the AAV candidate gene loci were extended by the closest gene(s) in the 5′ and 3′ direction, respectively, adding up to a total of 66 genes (HLA-DPA1 and HLA-DPB1 SNPs considered as one locus; supplementary Table S14, available at Rheumatology online). The complete sets of genome-wide significantly associated genes for stroke, coronary artery disease, peripheral artery disease and blood pressure were retrieved from the NHGRI-EBI GWAS Catalogue (supplementary Table S15, available at Rheumatology online) and analysed for the presence of any of the 66 candidate AAV genes. Genes in 7 out of 11 loci (64%) with (suggestive) association with PR3-ANCA+ AAV and 6 out of 10 MPO-ANCA+ AAV loci (60%) were significantly associated with CVD.
To determine whether there was an enrichment of CVD-associated loci among the 22 lead SNPs with an association/suggestive association with PR3/MPO-ANCA+ AAV, we used a simulation approach to estimate the NULL distribution for the frequency of overlap of randomly sampled sets of 22 SNPs (±100 000 bps centered on each SNP), from the total number of sequenced SNPs in the discovery cohorts, with CVD-associated SNPs (±100 000 bps centered on each SNP; NHGRI-EBI GWAS Catalogue; supplementary Table S15, available at Rheumatology online). The mean frequency of CVD overlap for the sampled SNP distribution was 0.27, compared with 0.45 for the observed CVD overlap for AAV SNP regions (P = 0.12; supplementary Fig. S6, available at Rheumatology online).
Discussion
In the present study, we used a large-scale candidate gene approach to identify disease-associated risk variants in AAV. The targeted sequencing study design allowed us to (i) enrich for candidate genes with a plausible impact on immune response; (ii) enrich for non-protein coding regions with a potentially regulatory function, by specifically selecting evolutionarily conserved sequences; and (iii) analyse common as well as rare genetic variants. As a result, we identified a novel association between a rare genetic variant at the BACH2 locus and MPO-ANCA+ AAV, and confirmed previous associations between AAV and the HLA and SERPINA1 loci.
The BACH2 SNP associated with MPO-ANCA+ AAV is rare, and hence not specifically tagged on traditional GWAS SNP arrays. Interestingly, another SNP at the BACH2 locus recently showed a suggestive association with EGPA, a subtype of AAV where a proportion of patients have MPO-ANCA [2]. In contrast, BACH2 has neither now nor previously been associated with PR3-ANCA+ AAV, supporting the notion that MPO-ANCA+ and PR3-ANCA+ AAV are separate entities with distinct genetic predisposition [3, 4].
In this study, there were no other SNPs at the BACH2 locus with a P-value of significant or suggested association with disease. According to our in vitro analyses, rs78275221 is located in an enhancer region functional in B cells and endothelium of dermal micro-vessels, where the risk allele eliminates the enhancing effect in endothelial cells. In silico predictions highlight RREB1 as a candidate transcription factor with its binding motif significantly affected by rs78275221. RREB1 may execute a repressive or activating effect on gene transcription [25]; hypothetically, in this case, RREB1 would function as a repressor when bound to the high-affinity rs78275221-A allele, resulting in increased gene expression in carriers of the low-affinity rs78275221-G wild-type allele.
Three genes are located within the TAD of rs78275221. One of them, BACH2, has been genetically associated with numerous autoimmune diseases and with the levels of various leukocytes [12–14, 26–30]. It is expressed in immune cells, endothelial cells and fibroblasts [31–33], and encodes a transcription factor essential for B cell maturation and T cell differentiation [34–36], where loss of function facilitates the development of autoimmunity [35, 37]. Collectively, our results suggest that in a subset of individuals with MPO-ANCA+ AAV, the development of vasculitis is favoured by reduced expression of a specific gene, possibly BACH2, in endothelial cells, orchestrated by a regulatory variant with cell type-specific function. Conceivably, it is the confinement to endothelium that connects this functional variant to vasculitis, rather than to other autoimmune disorders linked to the BACH2 locus. However, the effects of this variant in additional cell types and subsets, such as fibroblasts or T regulatory cells, have not been fully evaluated in this study and thus require functional follow-up studies.
While confirming the associations between AAV and the HLA region, using a re-sequencing approach we found that both common and rare (MAF >0.01) SNPs were significantly associated with PR3/MPO-ANCA. Numerous both low- and high-frequency variants had suggested coding or regulatory functions, generating a great number of combinations of putatively functional variants working in sets or as singlets. These results underscore the immense complexity in regulation and function of HLA molecules and the extraordinary heterogeneity among individuals.
A limitation of the current study is the relatively small sample sizes, particularly for MPO-ANCA+ AAV, hampering the statistical power to detect loci with significant association with AAV. Hence, the novel variant rs78275221 at the BACH2 locus did not fully reach the threshold for statistical significance in the MPO-ANCA+ discovery cohort of 175 cases, but did so in the meta-analysis of 276 cases. The limited statistical power leaves it open as to whether additional loci with suggestive association with disease in our discovery analysis are truly associated loci or false positives.
Interestingly, one of the loci with suggestive association with MPO-ANCA+ AAV in our study, HDAC9, is strongly associated with stroke and other manifestations of CVD [22–24, 38], as is the PR3-ANCA+ AAV-associated SERPINA1 locus [20]. Patients with AAV suffer an increased risk of cardio-vascular and thromboembolic events, largely attributable to the inflammation characterizing AAV [39–41]. The incidence rate of thromboembolism is, however, more than seven times higher in AAV than in other systemic autoimmune disorders, which, together with our preliminary findings, raises questions about a possible common denominator between AAV and CVD/thromboembolism [39].
In conclusion, in this study we expand the number of genetic loci that have been associated with AAV by identification of a novel susceptibility locus in the BACH2 gene for MPO-ANCA+ AAV. Our results suggest that a disease-associated variant at this locus is of mechanistic importance by exerting a regulatory function on gene expression in specific cell types.
Supplementary Material
Acknowledgements
Sequencing and MASSarray genotyping were performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computations and data handling were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX (Project SENS2017543), partially funded by the Swedish Research Council through grant agreement no. 2018-05973.
We thank the Biobank Research Unit at Umeå University, Västerbotten Intervention Programme, the Northern Sweden MONICA study and the County Council of Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council (no. 2017-00650).
Funding: The project was funded by Swedish Society for Medical Research, Swedish Research Council, Swedish Society of Medicine, Swedish Rheumatism Association and Knut and Alice Wallenberg Foundation (Scholar Award, KLT). Sequencing of a proportion of the controls was supported by an AstraZeneca-Science for Life Laboratory (SciLifeLab) Research Collaboration Grant, focussing on genetic studies of systemic lupus erythematosus, myositis and Sjögren’s syndrome (Dissect Consortium).
J.D. was supported by the Swedish Society for Medical Research, King Gustav V’s 80-year Foundation and Uppsala University Hospital. D.E. and B.S. were financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab. R.P. was funded by the Regional agreement on medical training and clinical research between the Western Götaland county council and the University of Gothenburg (RP, ALF/GBG-926621) and the Nanna Svartz Foundation. J.R.S.M. was supported by the Swedish Research Council, FORMAS (221-2012-1531).
Disclosure statement: The authors have declared no conflicts of interest.
Contributor Information
Johanna Dahlqvist, Department of Medical Sciences; Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden; Broad Institute of MIT and Harvard University, Cambridge, MA, USA.
Diana Ekman, Department of Biochemistry and Biophysics, National Bioinformatics Infrastructure Sweden, Science for Life Laboratory, Stockholm University, Stockholm.
Bengt Sennblad, Department of Cell and Molecular Biology, National Bioinformatics Infrastructure Sweden, Science for Life Laboratory, Uppsala University, Uppsala.
Sergey V Kozyrev, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Jessika Nordin, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Åsa Karlsson, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Jennifer R S Meadows, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Erik Hellbacher, Department of Medical Sciences.
Solbritt Rantapää-Dahlqvist, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Ewa Berglin, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Bernd Stegmayr, Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
Bo Baslund, Copenhagen Lupus and Vasculitis Clinic, Center for Rheumatology and Spine Diseases, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
Øyvind Palm, Department of Rheumatology, Oslo University Hospital.
Hilde Haukeland, Department of Rheumatology, Martina Hansens Hospital, Oslo, Norway.
Iva Gunnarsson, Department of Medicine, Division of Rheumatology, Karolinska Institutet, Stockholm; Unit of Rheumatology, Karolinska University Hospital, Stockholm.
Annette Bruchfeld, Department of Health, Medicine and Caring Sciences, Linköping University, Linköping; Department of Renal Medicine, Karolinska University Hospital and CLINTEC Karolinska Institutet, Stockholm.
Mårten Segelmark, Department of Clinical Sciences, Division of Nephrology, Lund University and Skåne University Hospital.
Sophie Ohlsson, Department of Clinical Sciences, Division of Nephrology, Lund University and Skåne University Hospital.
Aladdin J Mohammad, Department of Clinical Sciences Lund, Section of Rheumatology, Skåne University Hospital, Lund University, Lund, Sweden; Department of Medicine, University of Cambridge, Cambridge, UK.
Anna Svärd, Center for Clinical Research Dalarna, Uppsala University, Uppsala.
Rille Pullerits, Department of Rheumatology and Inflammation Research, Institution of Medicine, Sahlgrenska Academy at University of Gothenburg, Gothenburg; Department of Clinical Immunology and Transfusion Medicine, Sahlgrenska University Hospital.
Hans Herlitz, Department of Molecular and Clinical Medicine/Nephrology, Institute of Medicine, the Sahlgrenska Academy, University of Gothenburg, Gothenburg.
Annika Söderbergh, Department of Rheumatology, Örebro University Hospital, Örebro, Sweden.
Gerli Rosengren Pielberg, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Lina Hultin Rosenberg, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Matteo Bianchi, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Eva Murén, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden.
Roald Omdal, Clinical Immunology Unit, Department of Internal Medicine, Stavanger University Hospital, Stavanger; Department of Clinical Science.
Roland Jonsson, Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, Bergen, Norway.
Maija-Leena Eloranta, Department of Medical Sciences.
Lars Rönnblom, Department of Medical Sciences.
Peter Söderkvist, Department of Biomedical and Clinical Sciences, Division of Cell Biology.
Ann Knight, Department of Medical Sciences.
Per Eriksson, Department of Biomedical and Clinical Sciences, Division of Inflammation and Infection, Linköping University, Linköping, Sweden.
Kerstin Lindblad-Toh, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden; Broad Institute of MIT and Harvard University, Cambridge, MA, USA.
Data availability statement
The data underlying this article cannot be shared publicly as that would compromise research participant privacy and consent. The data will be shared on reasonable request to the corresponding author. Datasets located in the public domain can be derived from: ENCODE (www.encodeproject.org), HaploReg (pubs.broadinstitute.org/mammals/haploreg/haploreg.php), JASPAR (jaspar.genereg.net), Hi-C 3D interactions (http://3dgenome.fsm.northwestern.edu), LDlink (ldlink.nci.nih.gov/?tab=home), NHGRI-EBI GWAS Catalogue (www.ebi.ac.uk/gwas/), gnomAD (gnomad.broadinstitute.org), SweFreq (swefreq.nbis.se), Vascular single cell database (betsholtzlab.org/VascularSingleCells/database.html), GTEx (gtexportal.org).
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Kallenberg CG. Key advances in the clinical approach to ANCA-associated vasculitis. Nat Rev Rheumatol 2014;10:484–93. [DOI] [PubMed] [Google Scholar]
- 2. Lyons PA, Peters JE, Alberici F. et al. ; European Vasculitis Genetics Consortium. Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status. Nat Commun 2019;10:5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyons PA, Rayner TF, Trivedi S. et al. Genetically distinct subsets within ANCA-associated vasculitis. N Engl J Med 2012;367:214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Merkel PA, Xie G, Monach PA. et al. ; Vasculitis Clinical Research Consortium. Identification of functional and expression polymorphisms associated with risk for antineutrophil cytoplasmic autoantibody-associated vasculitis. Arthritis Rheumatol 2017;69:1054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willeke P, Schluter B, Sauerland C. et al. Farm exposure as a differential risk factor in ANCA-associated vasculitis. PLoS One 2015;10:e0137196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tam V, Patel N, Turcotte M. et al. Benefits and limitations of genome-wide association studies. Nat Rev Genet 2019;20:467–84. [DOI] [PubMed] [Google Scholar]
- 7. Watts R, Lane S, Hanslik T. et al. Development and validation of a consensus methodology for the classification of the ANCA-associated vasculitides and polyarteritis nodosa for epidemiological studies. Ann Rheum Dis 2007;66:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eriksson D, Bianchi M, Landegren N. et al. Extended exome sequencing identifies BACH2 as a novel major risk locus for Addison’s disease. J Intern Med 2016;280:595–608. [DOI] [PubMed] [Google Scholar]
- 9. Mägi R, Morris AP.. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karczewski KJ, Francioli LC, Tiao G. et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ameur A, Dahlberg J, Olason P. et al. SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet 2017;25:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Márquez A, Kerick M, Zhernakova A. et al. ; Type 1 Diabetes Genetics Consortium. Meta-analysis of Immunochip data of four autoimmune diseases reveals novel single-disease and cross-phenotype associations. Genome Med 2018;10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Onengut-Gumuscu S, Chen WM, Burren O. et al. ; Type 1 Diabetes Genetics Consortium. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. International Multiple Sclerosis Genetics Consortium (IMSGC)Beecham AH, Patsopoulos NA. et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013;45:1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fornes O, Castro-Mondragon JA, Khan A. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2020;48:D87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas-Chollier M, Hufton A, Heinig M. et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc 2011;6:1860–9. [DOI] [PubMed] [Google Scholar]
- 17. Rao SS, Huntley MH, Durand NC. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie G, Roshandel D, Sherva R. et al. Association of granulomatosis with polyangiitis (Wegener’s) with HLA-DPB104 and SEMA6A gene variants: evidence from genome-wide analysis. Arthritis Rheum 2013;65:2457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward LD, , Kellis M.. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malik R, Dau T, Gonik M. et al. ; International Stroke Genetics Consortium. Common coding variant in SERPINA1 increases the risk for large artery stroke. Proc Natl Acad Sci USA 2017;114:3613–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klarin D, Lynch J, Aragam K. et al. ; VA Million Veteran Program. Genome-wide association study of peripheral artery disease in the Million Veteran Program. Nat Med 2019;25:1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson CP, Goel A, Butterworth AS. et al. ; EPIC-CVD Consortium. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–91. [DOI] [PubMed] [Google Scholar]
- 23. Malhotra R, Mauer AC, Lino Cardenas CL. et al. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat Genet 2019;51:1580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Markus HS, Mäkelä KM, Bevan S. et al. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. Stroke 2013;44:1220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang S, Qian X, Redman C. et al. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene 2003;22:2285–95. [DOI] [PubMed] [Google Scholar]
- 26. Liu JZ, van Sommeren S, Huang H. et al. ; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Z, Guo Y, Shi H. et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol 2020;145:537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferreira MA, Mangino M, Brumme CJ. et al. ; International HIV Controllers Study. Quantitative trait loci for CD4: CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet 2010;86:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Astle WJ, Elding H, Jiang T. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 2016;167:1415–29.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen MH, Raffield LM, Mousas A. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 2020;182:1198–213.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He L, Vanlandewijck M, Mäe MA. et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci Data 2018;5:180160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vanlandewijck M, He L, Mäe MA. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018;554:475–80. [DOI] [PubMed] [Google Scholar]
- 33. Wang L, Yu P, Zhou B. et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat Cell Biol 2020;22:108–19. [DOI] [PubMed] [Google Scholar]
- 34. Muto A, Tashiro S, Nakajima O. et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 2004;429:566–71. [DOI] [PubMed] [Google Scholar]
- 35. Roychoudhuri R, Hirahara K, Mousavi K. et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 2013;498:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vahedi G, Kanno Y, Furumoto Y. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 2015;520:558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H, Hu Q, Zhang M. et al. Bach2 deficiency leads to spontaneous expansion of IL-4-producing T follicular helper cells and autoimmunity . Front Immunol 2019;10:2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duan L, Wei L, Tian Y. et al. Novel susceptibility loci for Moyamoya disease revealed by a genome-wide association study. Stroke 2018;49:11–8. [DOI] [PubMed] [Google Scholar]
- 39. Merkel PA, Lo GH, Holbrook JT. et al. ; Wegener’s Granulomatosis Etanercept Trial Research Group. Brief communication: high incidence of venous thrombotic events among patients with Wegener granulomatosis: the Wegener’s Clinical Occurrence of Thrombosis (WeCLOT) Study. Ann Intern Med 2005;142:620–6. [DOI] [PubMed] [Google Scholar]
- 40. Faurschou M, Mellemkjaer L, Sorensen IJ. et al. Increased morbidity from ischemic heart disease in patients with Wegener’s granulomatosis. Arthritis Rheum 2009;60:1187–92. [DOI] [PubMed] [Google Scholar]
- 41. Liapi M, Jayne D, Merkel PA, Segelmark M, Mohammad AJ.. Venous thromboembolism in ANCA-associated vasculitis. A population-based cohort study. Rheumatology (Oxford) 2021;60:4616–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly as that would compromise research participant privacy and consent. The data will be shared on reasonable request to the corresponding author. Datasets located in the public domain can be derived from: ENCODE (www.encodeproject.org), HaploReg (pubs.broadinstitute.org/mammals/haploreg/haploreg.php), JASPAR (jaspar.genereg.net), Hi-C 3D interactions (http://3dgenome.fsm.northwestern.edu), LDlink (ldlink.nci.nih.gov/?tab=home), NHGRI-EBI GWAS Catalogue (www.ebi.ac.uk/gwas/), gnomAD (gnomad.broadinstitute.org), SweFreq (swefreq.nbis.se), Vascular single cell database (betsholtzlab.org/VascularSingleCells/database.html), GTEx (gtexportal.org).