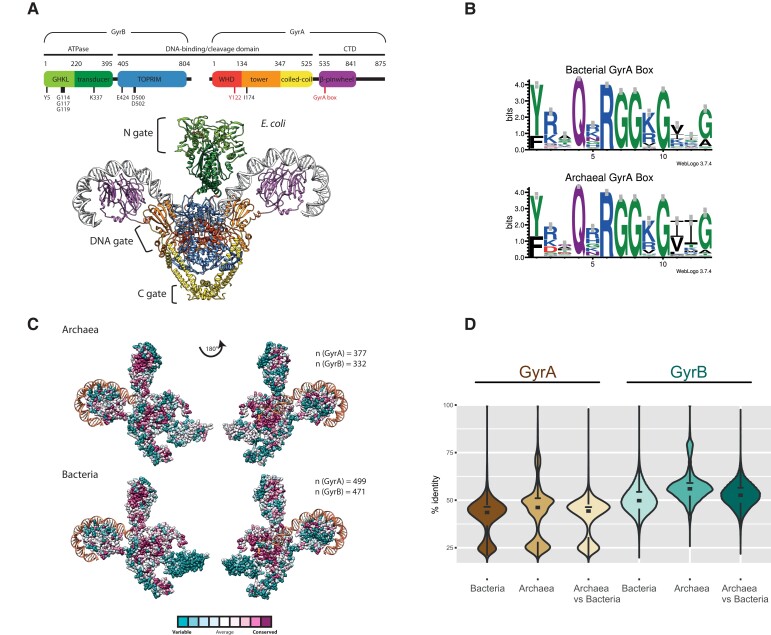
Fig. 1.
Sequence conservation of archaeal and bacterial DNA gyrases. (A) Structure of Escherichia coli DNA gyrase. A schematic representation of functional domains and catalytically important residues in GyrA and GyrB is shown. The GyrA box motif required for negative supercoiling activity and the catalytic tyrosine Tyr122 are highlighted in red. The three-dimensional structure of the A2B2 heterotetramer (PDB Nr. 6RKW) is shown bound to a 130 bp DNA duplex (same color code as the above scheme). The DNA duplex is depicted in grey. Gate regions are indicated to the left. (B) Comparison of the GyrA box motif in DNA gyrase sequences. The motif was generated using an alignment of 499 bacterial and 377 archaeal representative sequences. The letter size is proportional to the frequency of occurrence of each letter in the alignment. WebLogo v. 3.7.4. was used to generate the sequence logo (Crooks et al. 2004). (C) The conservation scores of DNA gyrase amino acids were computed by ConSurf server using the empirical Bayesian method from the multiple sequence alignment. The number of aligned sequences is indicated. The ConSurf conservation score was projected onto the structure of E. coli DNA gyrase (PDB 6RKW). For clarity, only one A and one B subunits are shown.(D) Sequence conservation analysis using pairwise BLAST. Sequence identity was determined using all against all BLASTp searches. The statistical analysis and graphical representation were generated using R packages (see Materials and Methods). The analysis shows that the mean level and distribution of sequence conservation within each domain or between the two domains is similar.