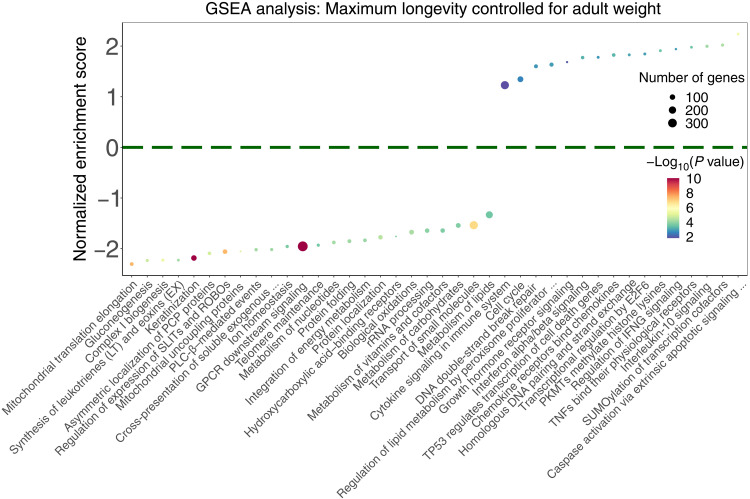
Fig. 1. Summary of the top significantly enriched pathways (adjusted P < 0.1) by the genes whose conservation scores are correlated with cancer resistance estimates (MLCAW), using GSEA with gene set annotations from the Reactome database.
The cancer resistance estimate used is “maximum longevity controlled for adult weight” (MLCAW). Normalized enrichment score is plotted on the y axis, where positive values correspond to enrichment by the PC genes and negative values correspond to enrichment by the NC genes. The dot color represents the significance of the enrichment (negative log10 GSEA P value), and the dot size represents the number of genes in the “leading edge,” i.e., the set of genes that are enriched in a pathway. For the sake of clarity, only a subset of the enriched pathways (FDR < 0.1) are shown, and long pathway names have been shortened (using “…”). The complete pathway enrichment results are given in table S3B.