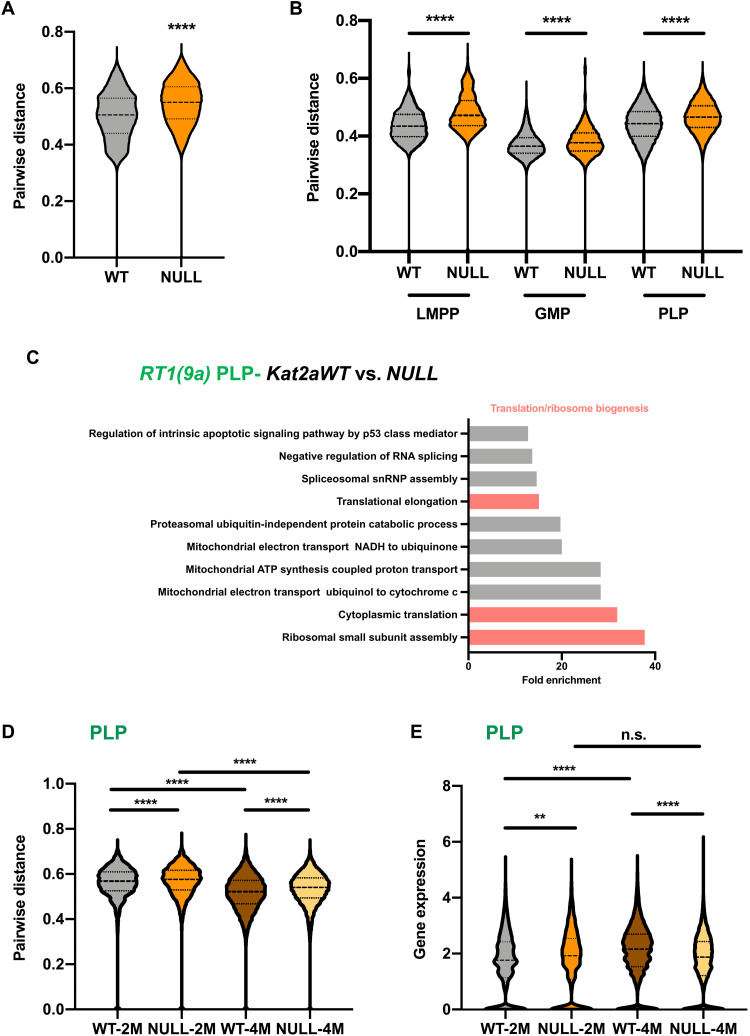
Fig. 5. Loss of Kat2a destabilizes expression of ribosomal biogenesis and translation-associated genes.
(A) Pairwise distance transcriptional variability measure (27) of Kat2aWT and Kat2aNULL RT1(9a) cells; top 500 most variable genes/genotype calculated by distance to the median CV (DM). ****P < 0.0001, nonparametric Kolmogorov-Smirnov (KS) test of cumulative distributions. (B) Comparison of RT1(9a) Kat2aWT and Kat2aNULL genotype-specific pairwise distances within individual LMPP, GMP, and PLP compartments. NULL up in all comparisons, with ****P < 0.0001, KS test. (C) Overrepresented gene ontology categories for genes down-regulated in RT1(9a) Kat2aNULL versus Kat2aWT PLP. *P-adj < 0.05. snRNP, small nuclear ribonucleoprotein; NADH, reduced form of nicotinamide adenine dinucleotide; ATP, adenosine 5′-triphosphate. (D) Pairwise distance of RT1(9a) PLPs. Comparisons consider correlations between ribosomal biogenesis genes, ****P < 0.0001, KS test. NULL up in all comparisons with WT; 2 months up in comparison with 4 months. (E) Distribution of expression levels for gene signatures in (C). **P-adj < 0.01, ****P-adj < 0.0001, two-tailed t test; n.s., not significant. NULL up at 2 months and down at 4 months in comparison with WT; WT up at 4 months relative to 2 months; comparison of NULL time points not statistically significant.