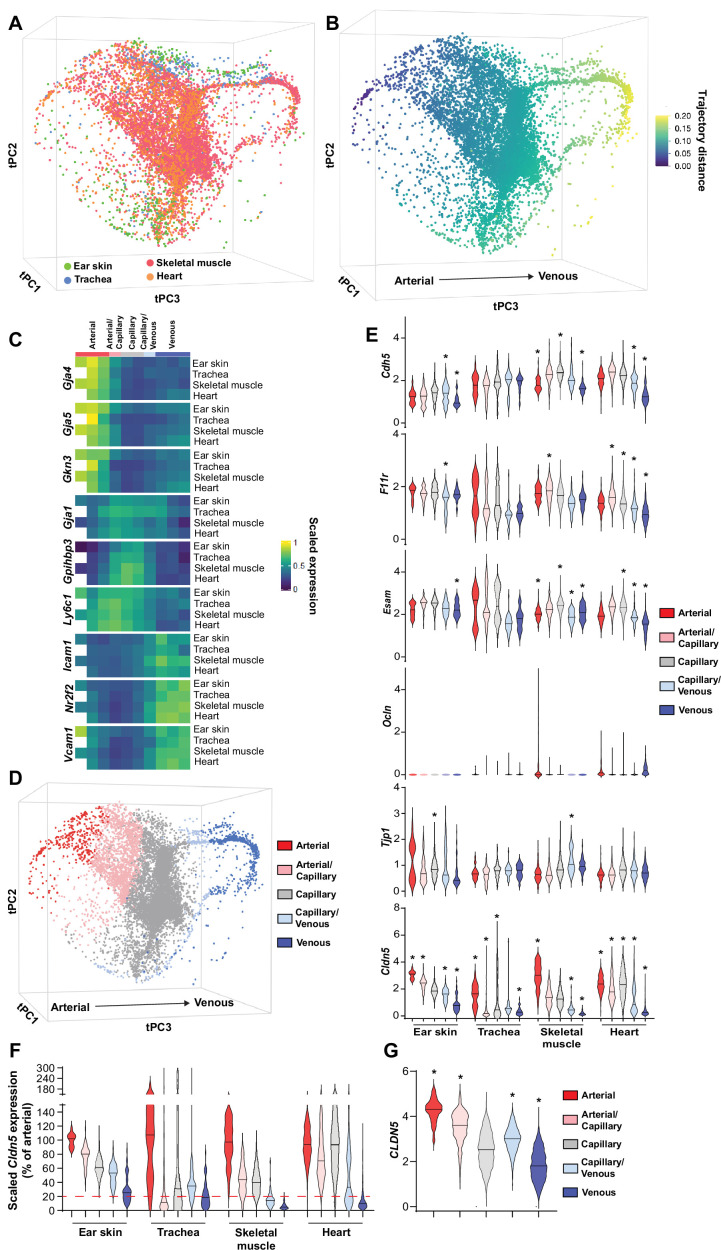
Figure 1. Patterning of the EC barrier at the single-cell level.
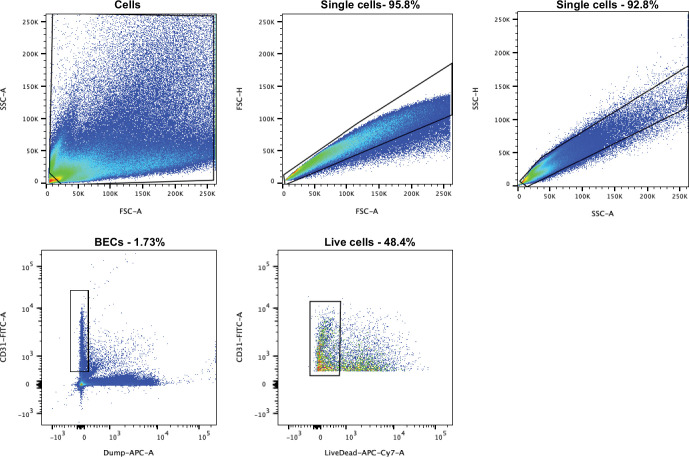
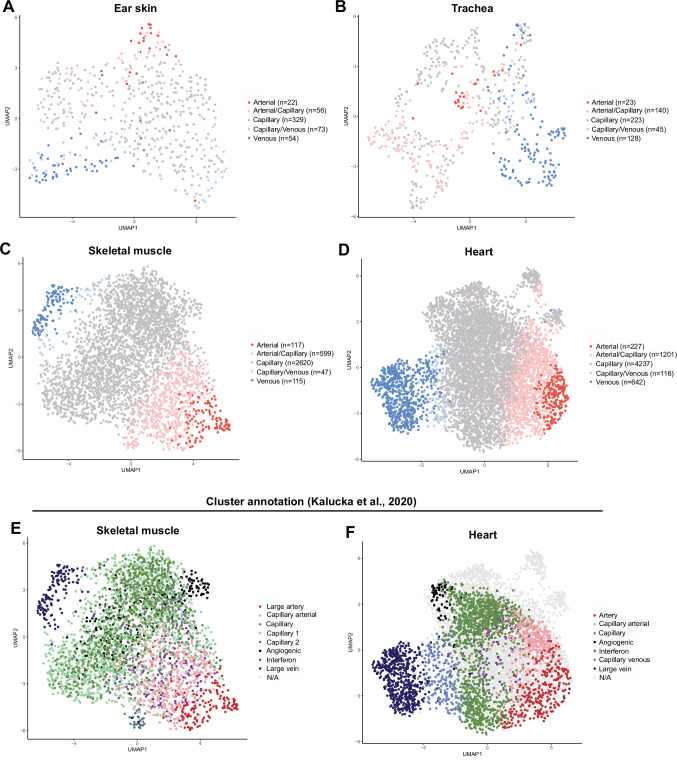
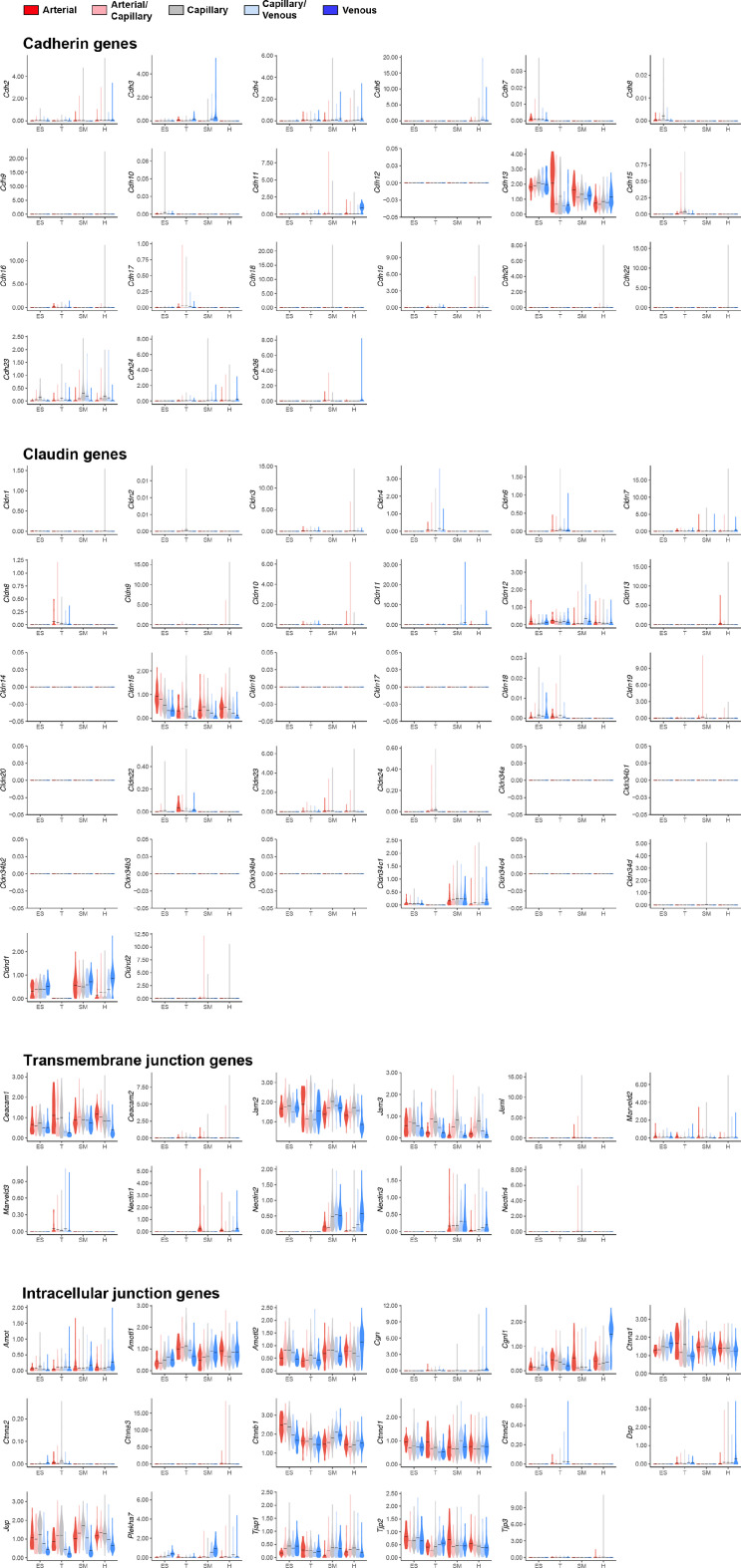
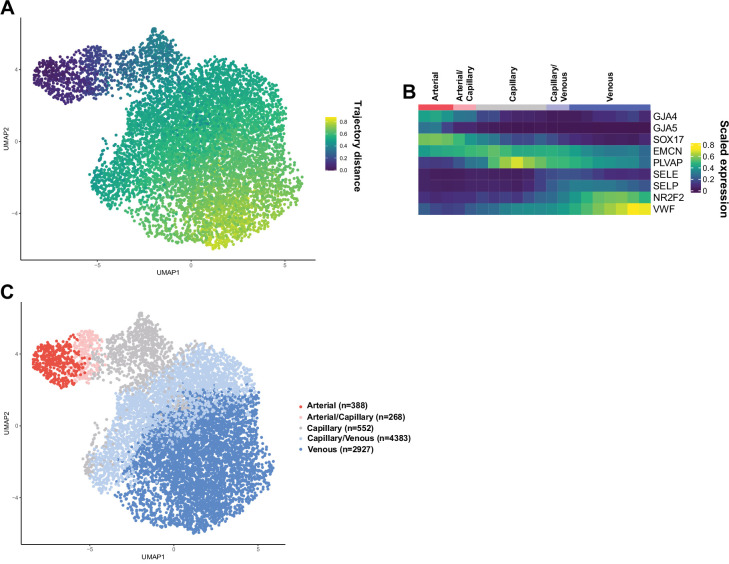
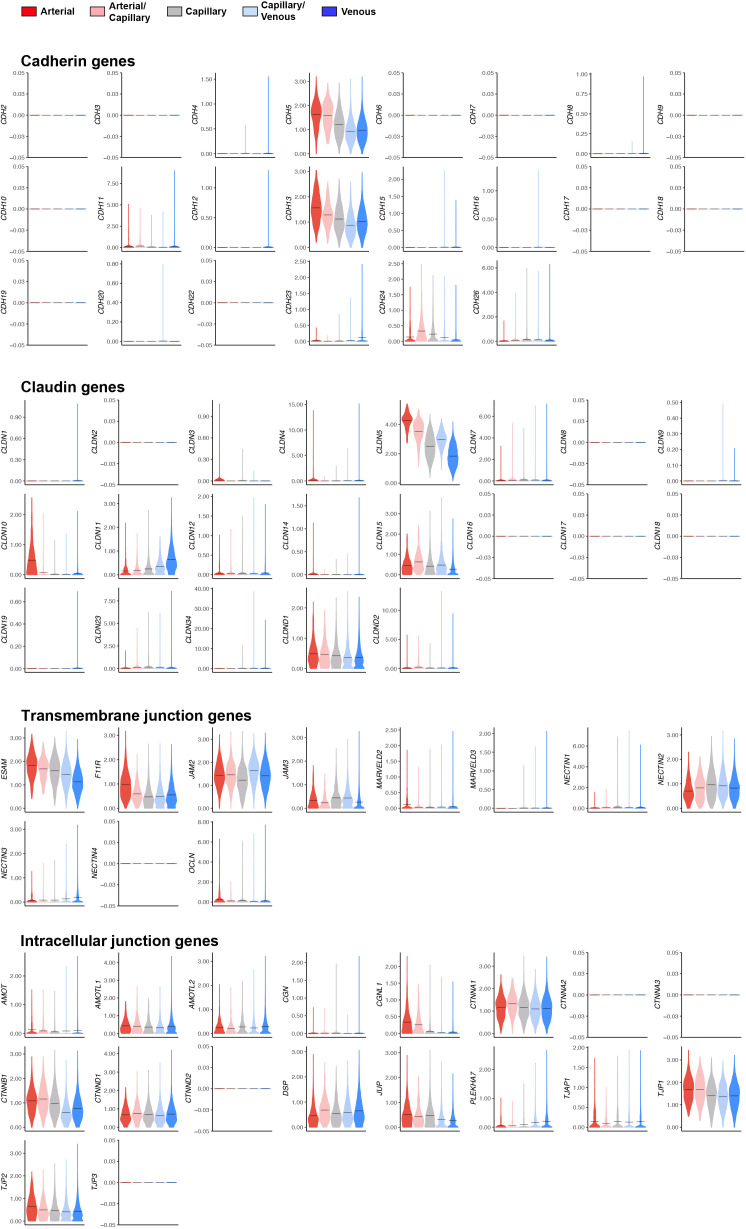
(A) Principal component analysis of the distances within 400 trajectories calculated with integrated data of murine datasets of ear skin, trachea, skeletal muscle, and heart blood endothelial cells (BECs). Colors illustrate the distribution of BECs (CD31+/CD45-/Lyve1-) for each organ. (B) Principal component analysis of trajectory distances colored by the distance along an isolated trajectory spanning from arterial to venous BEC. (C) Mean gene expression for each organ after equidistant binning of the isolated trajectory shown in B. Supervised vessel subset specifications (Top) based on the expression of previously established marker genes. (D) Principal component analysis of trajectory distances colored by the vessel subsets defined in C. (E) Violin plots of gene expression for BEC junctional components. Gene expression was normalized to account for differences in sample library size and has been imputed to account for dropouts in the data as described in Materials and methods. (F) Cldn5 expression in murine BEC datasets scaled per organ according to the mean expression in the arterial BECs of each organ. Red dashed line represents a fivefold reduction in expression compared to arterial BECs. (G) CLDN5 expression in human dermal BECs. n=534 ear skin, 559 trachea, 3498 skeletal muscle, 6423 heart and 8518 human BEC. * denotes statistical significance following differential gene expression analysis (Figure 1—source data 1–5).