Figure 3 |. Impact of SARS-CoV-2 VOC on neutralization capacity of vaccinated participants.
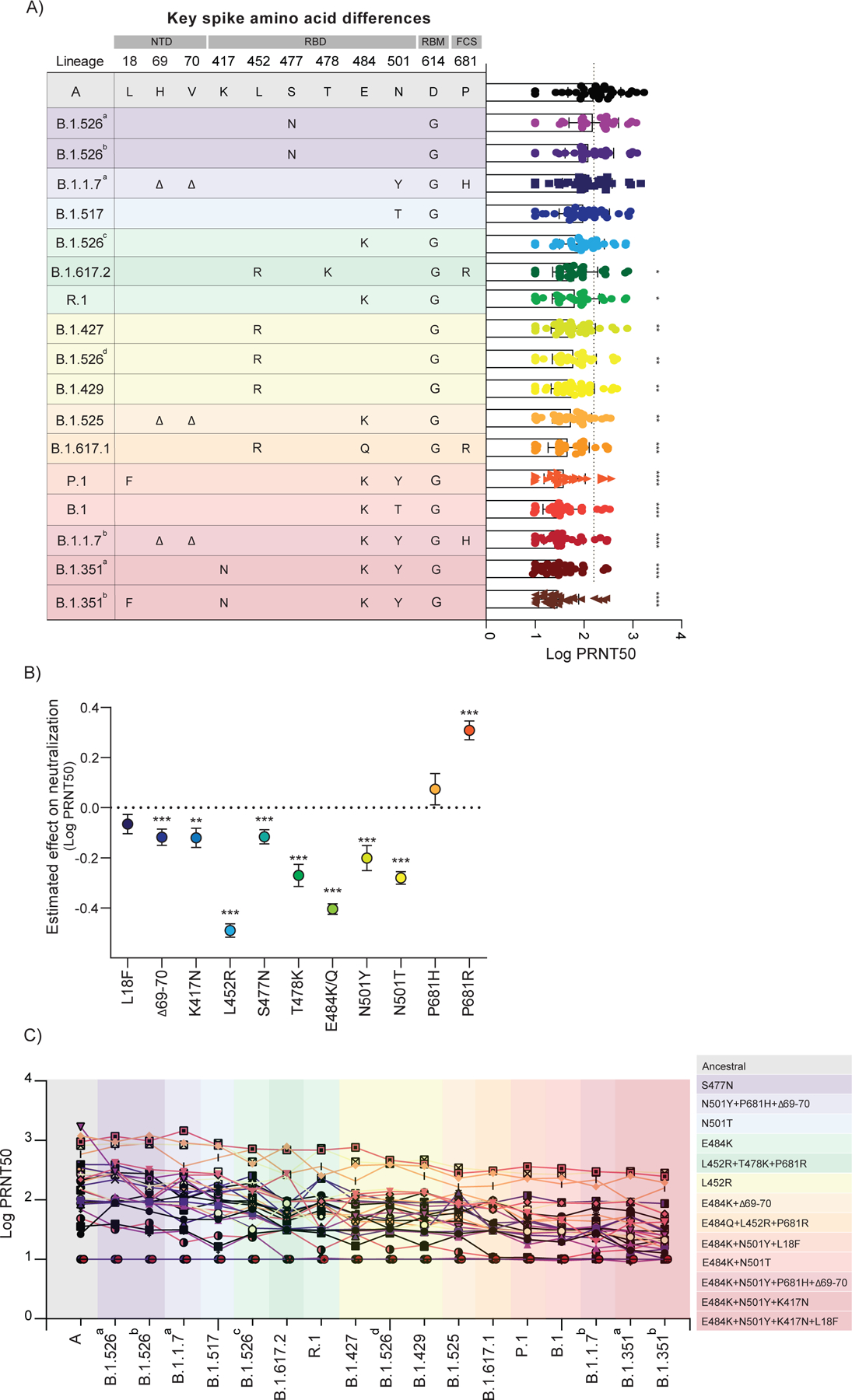
a, Plasma neutralization titers against ancestral lineage A virus, (WA1, USA) and locally circulating variants. Sixteen SARS-CoV-2 variants were isolated from nasopharyngeal swabs of infected individuals and an additional B.1.351 isolate was obtained from BEI. Neutralization capacity was assessed using plasma samples from vaccinated participants, 28 days post SARS-CoV-2 second vaccination dose at the experimental sixfold serial dilutions (from 1:3 to 1:2430). a, Key spike mutations within the distinct lineages and plasma neutralization titers (PRNT50). Spike mutations are arranged across columns and rows represent lineages. Significance was assessed by One-way ANOVA corrected for multiple comparisons using Dunnett’s method. Neutralization capacity of variants was compared to neutralization capacity against the ancestral strain. Boxes represent mean values ± standard deviations. Dotted line indicates the mean value of PRNT50 to ancestral strain. n=32. b, Estimated effect of individual mutations on plasma neutralization titers. Neutralization estimates (Log PRNT50) and significance were tested with a linear mixed model with subject-level random effects. Dots represent the ratio of linear mixed model coefficients + intercept to the intercept alone, and error bars represent the standard error. ****p < .0001***p < .001 **p < .01*p < .05. c, Individual trajectories of plasma neutralization titers (PRNT50). Each line represents a single individual. n=32. Dotted line indicates the mean value of PRNT50 to ancestral strain of previously infected individuals, prior to vaccination (baseline). Variants were grouped giving specific-spike mutations and are coloured accordingly. ****p < .0001 ***p < .001 **p < .01*p < .05. NTD, amino-terminal domain. RBD, receptor-binding domain. RBM, receptor-binding motif. FCS, furin cleavage site.
