Extended Data Figure 3 |. Maximum likelihood phylogeny of SARS-CoV-2 genomes of cultured virus isolates.
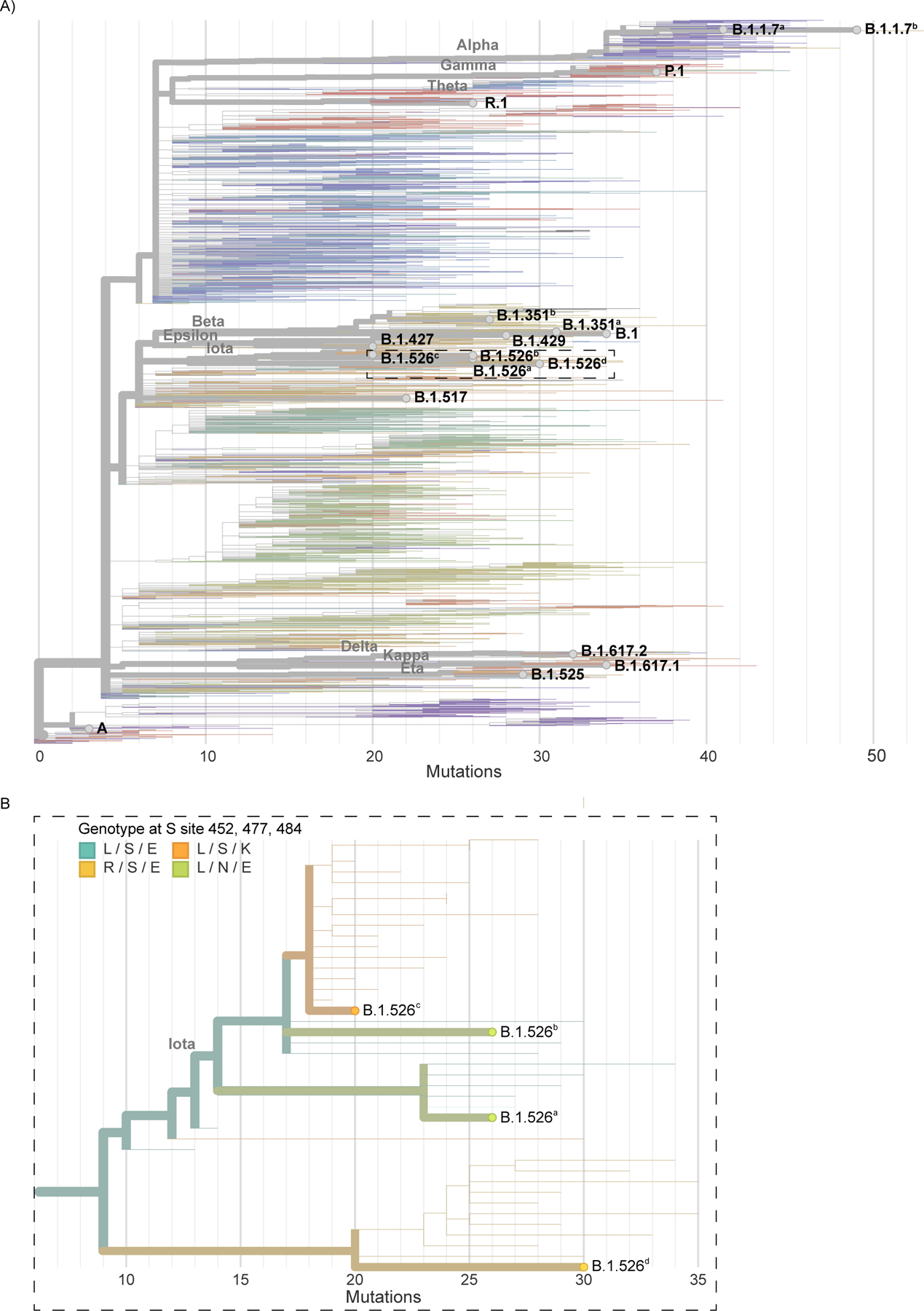
a, Nextclade (https://clades.nextstrain.org/) was used to generate a phylogenetic tree to show evolutionary relations between the cultured virus isolates used in this study and other publicly available SARS-CoV-2 genomes. Branches are colored by Pango Lineage, and labelled according to the WHO naming scheme. Highlighted are the cultured virus isolates used in this study. b, Enlarged section of the phylogenetic tree highlighting spike amino acid changes in the B.1.526 (iota) lineage viruses belonging to different clades.
