Abstract
The coronavirus known as COVID-19, which causes pandemics, is causing a global epidemic at a critical stage today. Furthermore, novel mutations in the SARS-CoV-2 spike protein have been discovered in an entirely new strain, impacting the clinical and epidemiological features of COVID-19. Variants of these viruses can increase the transmission in wastewater, lead to reinfection, and reduce immunity provided by monoclonal antibodies and vaccinations. According to the research, a large quantity of viral RNA was discovered in wastewater, suggesting that wastewater can be a crucial source of epidemiological data and health hazards. The purpose of this paper is to introduce a few basic concepts regarding wastewater surveillance as a starting point for comprehending COVID-19′s epidemiological aspects. Next, the observation of Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) in wastewater is discussed in detail. Secondly, the essential information for the initial, primary, and final treating sewage in SARS-CoV-2 is introduced. Following that, a thorough examination is provided to highlight the newly developed methods for eradicating SARS-CoV-2 using a combination of solar water disinfection (SODIS) and ultraviolet radiation A (UVA (315-400 nm)), ultraviolet radiation B (UVB (280-315 nm)), and ultraviolet radiation C (UVC (100-280 nm)) processes. SARS-CoV-2 eradication requires high temperatures (above 56°C) and UVC. However, SODIS technologies are based on UVA and operate at cooler temperatures (less than 45°C). Hence, it is not appropriate for sewage treatment (or water consumption) to be conducted using SODIS methods in the current pandemic. Finally, SARS-CoV-2 may be discovered in sewage utilizing the wastewater-based epidemiology (WBE) monitoring method.
Keywords: SARS-CoV-2, Wastewater-based epidemiology, UV, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Omicron (B.1.1.529)
Graphical abstract
1. Introduction
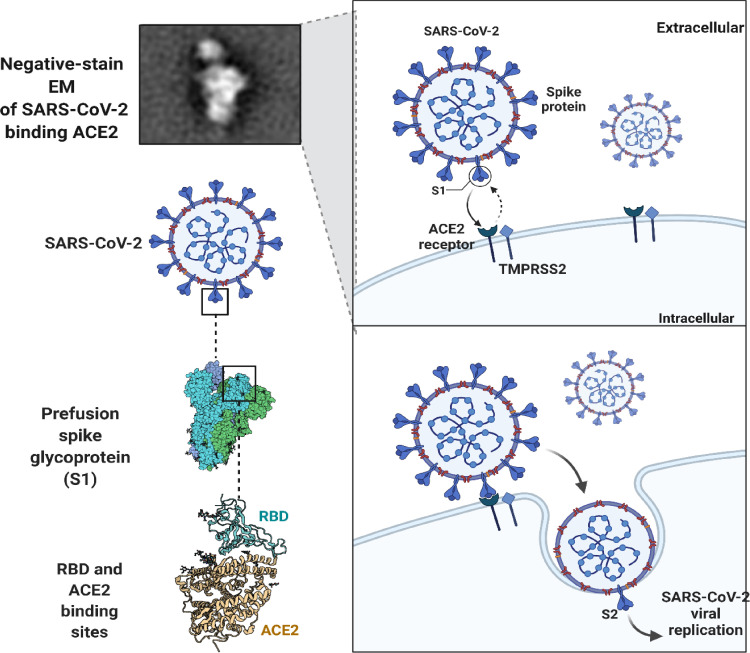
An outbreak of COVID-19 occurred in December 2019 and the World Health Organization declared it a worldwide threat in March 2020. Several single-stranded RNA viruses are associated with respiratory disease in humans (Pal et al., 2020). SARS-CoV-2 infections can result in respiratory or gastrointestinal problems when viral spike proteins bind to human angiotensin-converting enzyme 2 (ACE-2) receptors, causing respiratory or gastrointestinal problems. Infections caused by SARS-CoV-2 are similar to those caused by MERS, SARS-CoV, and other Betacoronaviruses ( Fig. 1 ) (Zhou et al., 2020). A change in the genetic sequence of the virus caused numerous variants of SARS-CoV-2 to arise during the epidemic.
Fig. 1.
SARS-CoV-2 particle targets ACE2 receptors in order to enter infected cells.
On the other hand, compared with other variations, most include minor changes, whereas specific acquired mutations give exceptional fitness by boosting transmissibility and resisting medical interventions, such as vaccinations. These variants have been associated with an increased risk; thus, they are deemed of concern (Harvey et al., 2021). A total of five variations have been categorized as a concern. In October 2020, the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) varieties appeared in the United Kingdom, South Africa, Brazil, and India, becoming the dominant strains in mid-2021 (Bugembe et al., 2021). In November 2021, five virulent concerns were reported across multiple countries, including Omicron (B.1.1.529, South Africa). The Omicron variant is notable for having 26_32 mutations in the spike protein. In spite of early results showing that Omicron is more infectious and immune evading than Delta, public health authorities are concerned about its rapid spread throughout South Africa and worldwide. In addition, only 10% of COVID-19 cases contain the Omicron variant (Pulliam et al., 2021).
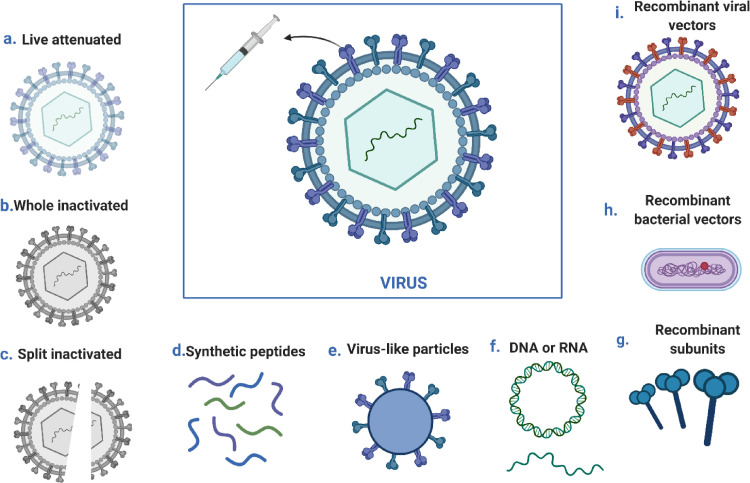
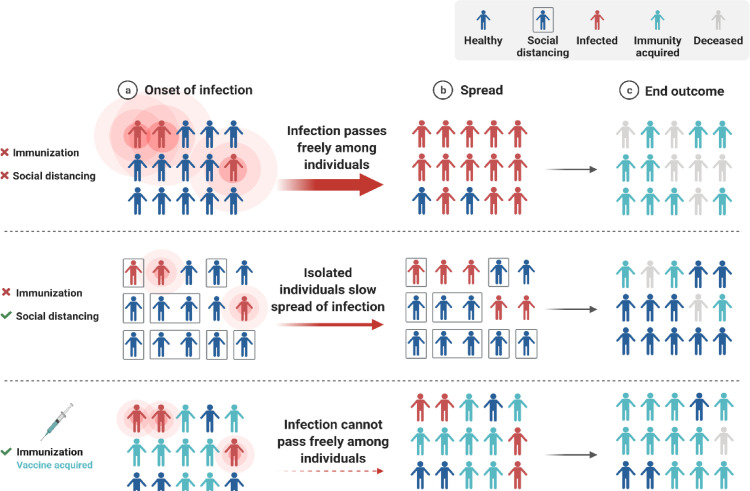
Public health remains seriously impacted by the COVID-19 pandemic despite ongoing threats (Zahmatkesh et al., 2022). In addition to protecting individuals, vaccination programs are helping society to open up and return to normalcy (Fig. 2 ) (Vissers et al., 2021). Nonetheless, vaccination strategies are at risk if some SARS-CoV-2 variants can evade the immune system (Fig. 3 ) (Dubey et al., 2022). The efficacy of the SARS-CoV-2 vaccination can only be retained by developing a robust, efficient, and proactive monitoring approach for new or known variations (Polo et al., 2020, Thompson, 2020). In addition to enhancing public health response and facilitating variants of concern's (VOCs') containment, molecular diagnostics provides a faster and more comprehensive diagnosis than genomic sequencing alone.
Fig. 2.
A schematic illustration of the development methods for viral vaccines; presenting a live attenuated vaccine, a whole inactivated vaccine, a split inactivated vaccine, a synthetic peptide vaccine, a virus-like vaccine, a DNA vaccine, a DNA vaccine, a bacterial vaccine, an RNA vaccine, a recombinant vector, and a virus vaccine (Bilal et al., 2020).
Fig. 3.
A theory of herd immunity and a concept of social distance.
Genomic sequencing of clinical samples currently serves as the basis for diverse surveillance. A downside of genomic sequencing is that it is neither accessible nor sustainable because of its high cost and specialized infrastructure. Sequencing is necessary for a meaningful dataset for a significant number of confirmed cases. Moreover, monitoring the spread and introduction of these new threats in flavivirus-naive and vaccinated populations is becoming increasingly essential to combating the pandemic. This pandemic has prompted much interest in wastewater-based surveillance (WBS). WBS provides a comprehensive, real-time view of the population in the catchment area at a very affordable price. Additionally, this method provided a straightforward way to determine the trends across different countries during the COVID-19 pandemic (Crits-Christoph et al., 2021; Fontenele et al., 2021; Napit et al., 2021).
Furthermore, the WBS technique has the potential to track the spread of SARS-CoV-2 strains in wastewater. Monitoring wastewater change is typically achieved through enriching and sequencing SARS-CoV-2 genomes that are detected in wastewater. Unfortunately, the lack of quantitative data modeling and low sensitivity to low-frequency variants limits its widespread application. In clinical samples, the capacity to target particular variations of the SARS-CoV-2 genome has been established utilizing RT-qPCR technologies (Van Poelvoorde et al., 2021). Measurement of multiple variants of wastewater has been adapted and validated with these techniques (Graber et al., 2021; Yaniv et al., 2021). In contrast to sequence-based methods, RT-qPCR is more sensitive and this technology distinguishes specific variant-linked mutations; thus, results can be quantified and interpreted within hours (Lee et al., 2021).
In September 2020, patients began acquiring new types of SARS-CoV-2 virus. Amongst them, the Alpha mutation (B.1.1.7) and Beta (B.1.351) are two of several genetically-altered variants. They are mainly resistant to neutralization by serum and plasma antibodies derived from convalescent patients (Trimpert et al., 2021). For the purposes of controlling the pandemic, community-level surveillance is necessary to identify potential public health issues (Mao et al., 2020). Thus, S proteins determine whether a virus is infective or transmissible (Syed et al., 2021). SARS-CoV-2′s receptor-binding domain (RBD) contains six essential amino acids critical for virus entry into host cells (L455, F486, Q493, S494, N501, and Y505) (Kim et al., 2021). Despite the link between spike protein (S) and human ACE2, polymorphisms in other amino acids may alter viral infection. There is an urgent need to understand the effects of amino acid substitutions in S proteins from new variants compared to the original strain on the transmissibility and virulence of SARS-CoV-2 (Mehra et al. 2021; Nagy et al., 2021; Xia, 2021).
As of November 26, 2021, the Omicron version of B.1.1.529 COVID-19 has been identified (COVID et al. 2021). In response to reports that the disease is more infectious and may be immune evading, flights were banned, and more measures were taken to control its spread in various regions of the world (Chen et al., 2021; Gu et al., 2021). Besides community-based monitoring of SARS-CoV-2 mutations, wastewater surveillance has been demonstrated to be a proper supplemental strategy (Hrudey et al. 2021; Lee et al., 2021). An epidemiological study retrospectively on routine surveillance data revealed that Omicron viruses might increase the risk of reinfection following primary infection (Ferré et al., 2022). A study in Nature suggests that omicron may be capable of evading immunity, especially for SARS-CoV-2 infections found among individuals with confirmed laboratory diagnoses. Over 2,796,982 individuals, 35,670 suspected reinfections were detected (Pulliam et al., 2021). As new variants are released, the severity of COVID-19 may increase or decrease. Variable factors impact COVID-19 severity and mortality, such as vaccination coverage, population demographic characteristics, age, socioeconomic status, comorbidities, management guidelines, and the simultaneous presence of numerous cases saturating the health system. Controlling these parameters allows large-scale case-control studies to establish clinical severity (Lee et al., 2021).
This paper argues that a wastewater treatment facility is necessary for the treatment of water generated by private homes, public facilities, and industrial facilities. Through the "sewage system", this water is taken to facilities that treat it through the " sewage system", before being discharged into the environment. To control and surveil infectious diseases, monitoring wastewater and WBEs is crucial. Among the essential aspects of combating SARS-CoV-2 are prevention and control within the community and continuous surveillance. By monitoring RNA in genetic material and screening for virus presence in the entire community, sewage or wastewater that contains viruses like SARS-CoV-2 has the capability to be detected. In addition, SARS-CoV-2 can be shed through stools; thus, wastewater is likely to contain the virus.
2. SARS-CoV-2′s practical application in wastewater treatment
There was the first detection of Alphas (B.1.1.7), Betas (B.1.351), Gammas (P.1), Deltas (B.1.617.2), and Omicrons (B.1.1.529) by a wastewater surveillance program. As a result, variant tracking data cannot be used to confirm the existence of a specific variant because all of the mutations required to define a variant cannot be determined on a single genome (Zahmatkesh et al., 2022, Zahmatkesh and Sillanpää, 2022). The presence of multiple mutations associated with variants, as well as mutations that are linked (i.e., mutations on a single sequence read), is more likely to increase confidence in the results. Identifying unique mutations not shared by known variants, collecting data on RNA concentration corresponding to emergence (for example, a low initial concentration followed by an increase over time), and detecting multiple variant-associated mutations trending together in consecutive samples or using multiple methods are all important (Zahmatkesh et al., 2022, Zahmatkesh and Sillanpää, 2022). Clinical case reports in the area should also be considered as well. There are several limitations to variant tracking in wastewater, such as detections that do not align with the current epidemiology, low quality sequencing data, sporadic detections, detections associated with a single variant, and conflicting trends in concentrations or abundances associated with the same variant. The usefulness of this data is limited by reporting times longer than one week (Zahmatkesh et al., 2022, Zahmatkesh and Sillanpää, 2022).
Detection of mutants associated with all mutants in community wastewater indicated that mutations associated with all mutants were more abundant than previously reported by clinical testing alone; all mutations associated with mutants were recorded. In order to detect emerging variants early, wastewater sampling can be used in conjunction with clinical testing. A public health messaging strategy and testing strategies can be derived from this information to guide decisions about the allocation of clinical and public health resources (Zahmatkesh et al., 2022, Zahmatkesh et al., 2022, Zahmatkesh and Sillanpää, 2022).
3. Evaluating the epidemiology of COVID-19 by monitoring wastewater
WBE has proven to be one of the significant successful methods of tracking viral circulation in a community, providing data on the frequency, variations in genetic profile, and distributions geographically of viruses within that group (Sinclair et al., 2008; Xagoraraki et al. 2020). Viruses excreted in feces can be identified by the general wastewater analysis from a specific area (Carducci et al., 2006; La Rosa et al. 2013). As a result, this method allows monitoring viral infection epidemics even when clinical surveillance does not reveal the infection, mainly because many viral infections are asymptomatic and clinical cases are rarely diagnosed (Johansson et al., 2014; Qi et al., 2018). The restrictions are equally applied to viruses shed via feces, including adenoviruses, noroviruses, sapoviruses, enteroviruses, rotaviruses, and hepatitis a viruses (Okabayashi et al., 2008; Rodriguez-Lazaro et al., 2012). To viruses that surveillance systems rarely or never reported, including Saffold virus, cosavirus, and salivirus/klasseviruses (Bonanno Ferraro et al., 2020; Kitajima et al., 2015; Kitajima et al., 2014; Thongprachum et al., 2018).
According to various reports, SARS-CoV-2 infections have been described as asymptomatic or nonsymptomatic (Lai et al., 2020; Mizumoto et al., 2020; Nishiura et al., 2020); however, estimating the extent of viral circulation in a community can be challenging. In addition, comparing nations with varying diagnostic skills and procedures to ascertain the natural level of viral circulation, is difficult. Additionally, wastewater surveillance could provide in-depth information on infection outbreaks in different regions, even if the clinical diagnosis is impractical or unavailable, such as in countries with limited diagnostic resources. Phylogenetic analysis has also been demonstrated to be an effective method of detecting changes in strains over time through wastewater monitoring, providing the ability to compare strains across regions and evaluate the evolutionary origins of strains, as has been demonstrated in previous studies of enteric viruses and, more recently, of the SARS-CoV-2 virus (Bisseux et al., 2018; La Rosa et al., 2014; Nemudryi et al., 2020).
As part of wastewater observation, finding low levels of viruses can be of importance; this can be demonstrated by the fact that the number of infections decreased after public health interventions such as the eradication of the poliovirus. The method has the capability of not only determining a new virus outbreak within a population, but can also identify seasonal variations in precipitation and temperature (Asghar et al., 2014). The purposes of this surveillance approach include serving as an early warning indicator that SARS-CoV-2 has been reintroduced in a community (Savolainen-Kopra et al., 2011; Sinclair et al., 2008). Additionally, investigating the effectiveness of public health intervention methods for reducing exposures related to lockdowns, loneliness, and social disengagement following public health interventions could be of benefit (Hellmér et al., 2014; Prevost et al., 2015; Sedmak et al., 2003). In addition to detecting new viruses from wastewater, viral virome analyses can allow the dissemination of resources to areas that may be infected and allow preventative measures to be taken before they become clinically apparent (Xagoraraki et al. 2020). According to recent Monte Carlo simulations, an estimate of the number of copies of SARS-CoV-2 RNA that can be found in untreated wastewater and the number of individuals affected has been made. The results of the model indicate that 171 to 1090 individuals were infected, which is consistent with clinical findings, and is consistent with the results of the study (Ahmed et al., 2020).
The usefulness and accuracy of wastewater monitoring may be increased by more excellent methodological and molecular test validation. Although wastewater surveillance can be used for many purposes, there is a limit to its use. In terms of epidemiologically determined cases and viral levels, a correlation may be complex due to differences in viral excretion rates during infection. As well as factors such as the length of transmission time, and a lack of consistency in the collection of spatial variability that occurs when traveling and transporting wastewater between multiple systems and dilution caused by precipitation or inactivation throughout the transit procedure and/or frequent clinical testing. There is a possibility that detecting and quantifying viruses may be challenging due to factors such as stable DNA in wastewater, inefficient methods of virus concentration, sample variability (mostly grabbing samples as opposed to composing samples). Moreover, the absence of sensitive kits, exceptionally at low viral concentrations (La Rosa et al. 2013).
Despite these challenges, the objective of implementing environmental monitoring systems for SARS-CoV-2 has not been achieved. A number of countries, including the Netherlands, France, the United States, and Australia, have identified SARS-CoV-2 in wastewater (Ahmed et al., 2020; Lodder et al. 2020; Medema et al., 2020; Nemudryi et al., 2020; Wu et al., 2020; Wurtzer et al., 2020). An example of recent research performed in the USA utilized wastewater monitoring to discover the genetic origins of the SARS-CoV-2 virus and estimate the efficiency of social isolation as a public health strategy to limit the epidemic (Nemudryi et al., 2020). Recent research, mainly continuing throughout the globe, may provide essential data for assessing the incidence of SARS-CoV-2 in communities and notifying communities that may minimize disease transmission. Public acceptability of wastewater monitoring may necessitate stressing ethical considerations linked to cleanliness, privacy, and rights. The WBE may give epidemiological statistics regarding illness prevalence in a community, thereby avoiding stigmatization associated with clinical diagnosis in epidemics like COVID-19 (Murakami et al., 2020).
3.1. Determination of Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) in wastewater
According to recent reports, SARS-CoV-2 RNA was found at the same level as other enteric viruses in the feces of COVID-19 patients. Despite the fact that SARS-CoV-2 has the potential to be identified in untreated sewage, as was the case previously, a viral concentration step may still be necessary in order to detect the virus (Medema et al., 2020; Prüss et al., 2002; Wurtzer et al., 2020; Wyn‐Jones et al. 2001).
As a model virus, most studies have been conducted on viruses that can be cultured or on bacteriophages, which include enveloped as well as non-enveloped enteric viruses such as norovirus, enterovirus, adenovirus, and hepatitis (Haramoto et al., 2018). Several methods of concentrating enteric viruses in water samples, whether treated or untreated, have been developed based on membranes with electropositive and electronegative properties (Cashdollar et al. 2013; Haramoto et al., 2018). The process of enteric virus infection involves electrostatic interactions between filters and enteric viruses. When in water with almost neutral pH, a significant number of enteric viruses are electrostatically negatively charged. A positively charged virus particle can be bind directly to an electropositive filter or indirectly to an electronegative filter by virtue of salt-bridging (Hill et al., 2005; Ikner et al., 2012; Michen et al. 2010). Viral concentrations can be achieved using ultrafiltration, a process that utilizes the principle of size exclusion to concentrate bacteria from ambient water samples. Other techniques have been employed to isolate viruses in wastewater samples, such as polyethylene glycol (PEG) (Lewis et al. 1988), ultracentrifugation (Fumian et al., 2010), and the use of skimmed-milk flocculation (Calgua et al., 2013).
A variety of indigenous enteric viruses have been successfully detected using these virus concentration methods without using any model viruses during their development (Fong et al. 2005; Haramoto et al., 2018). Due to the limits of existing viral concentration technologies, limited information is known on the recovery efficiency of encapsulated viruses, including CoVs. According to Ye et al., and Wigginton (2016), enveloped viruses such as mouse hepatitis virus (MHV) and Pseudomonas phage Φ6 adsorb more to the solid part of sewage than nonenveloped viruses (Ye et al., 2016). Haramoto et al., and Ohgaki (2009) report that enveloped koi herpesviruses are able to adhere to an electronegative filter with an average absorption rate of 87% or higher (Haramoto et al., 2009). These findings suggest that filter filtering may be an effective technique for recovering SARS-CoV-2 from water and sewage; however, more research is necessary. As a result of the virus and the aquatic environment, virus recovery behavior can vary greatly, even within enteric viruses (Haramoto et al., 2018). Therefore, due to a difference between SARS-CoV-2 and enteric viruses in structure and physical properties, this method of virus concentration is not very useful for evaluating its efficacy. For instance, an electropositive membrane filter method, used by Wang et al., did not recover SARS-CoV from wastewater, a significant decrease in recovery compared to other types of enteroviruses (Wang et al., 2005).
However, SARS-CoV-2 concentrations in wastewater will most certainly need to be more significant for detection, and further research should be conducted to determine the effectiveness of the recovery process. At the same time, further research must be conducted in order to determine how effective these existing methods are for concentrating on SARS-CoV-2. Due to their low pathogenicity, MHV and human CoV strains may be utilized as models to assess and improve SARS-CoV-2 techniques. Several elements potentially influence the development of SARS-CoV-2 models, including Pseudomonas phage Φ6. SARS-CoV-2 RNA analysis in wastewater has been conducted using a variety of concentration methods in Australia, France, the Netherlands, and the United States, including ultrafiltration, precipitation with PEG, electronegative membrane adsorption, and direct RNA extraction (Wu et al., 2020; Wurtzer et al., 2020).
The water concentration considerably impacts the identification of viruses in the sample; for example, untreated sewage samples should be concentrated at 100 mL in order to identify enteric viruses (Haramoto et al., 2018). For preliminary studies, rapid molecular detection of SARS-CoV-2 was accomplished by evaluating up to 200 mL of untreated sewage specimens. However, if SARS-CoV-2 is less widespread in the local area, processing a more significant number of wastewater samples may be appropriate (Medema et al., 2020; Nemudryi et al., 2020).
A novel testing method has been developed to detect SARS-CoV-2 using RT-qPCR and nested RT-PCR (Corman et al., 2020; Coronavirus, 2019; Organization, 2020; Shirato et al., 2020). These methods were employed to detect SARS-CoV-2 in the new testing approach. Assays based on TaqMan have been developed by Corman et al. (2020) for detecting three genes: RNA-dependent RNA polymerase (RdRp), envelope (E), and nucleocapsid (N) with detection limits of 3.8, 5.2, and 8.3 RNA copies per reaction for three genes. It has been explained above that SARS-CoV-2 is identified using RT-qPCR using the RdRp sequence ( Table 1 ). After this, the SARS-CoV-2 is compared to the SARS-CoV-2 and bat-SARS-related CoVs, while the SARS-CoV-2 and SARS-CoV-2 are detected by E gene-RT-qPCR. Due to its significantly larger ALOD than the two others, the performance of the N gene-RT-qPCR test was not thoroughly examined in the study (Fig. 4 ) (Corman et al., 2020). In contrast, Shirato et al. (2020) observed that only RT-qPCR of N genes recognized SARS-CoV-2 and performed well on their RT-qPCR platform when compared to the three other tests. The N protein gene is the most extensively used as a candidate for RT-qPCR assays (Shirato et al., 2020). According to Shirato et al. (2020), RT-qPCR can identify as few as five copies of RNA per reaction.
Table 1.
Human feces contain SARS-CoV-2.
| Process of detection | SARS-CoV-2 positive patients' rate | Infection rate (copies/L) | References |
|---|---|---|---|
| rRT-PCR | 44/153 (29%) | < 2.6 × 107 | (Wang et al., 2020) |
| Cell-culture | 2/4 (50%) | - | (Wang et al., 2020) |
| rRT-PCR | 31/65 (48%) | - | (Lin et al., 2020) |
| rRT-PCR | 28/42 (67%) | - | (Chen et al., 2020) |
| - | 9/59 (15%) | 1.26 × 108 (with diarrhea) | (Cheung et al., 2020) |
| rRT-PCR | 39/73 (53%) | - | (Xiao et al., 2020) |
| RT-qPCR | 10/10 (100%) | - | (Lo et al., 2020) |
| rRT-PCR | 5/14 (36%) | - | (Zhang et al., 2020) |
| rRT-PCR | 5/13 (38%) | - | (Li et al., 2020) |
| rRT-PCR | 41/74 (55%) | - | (Wu et al., 2020) |
| rRT-PCR | 9/17 (53%) | 5.5 × 104–1.21 × 105 | (Pan et al., 2020) |
| rRT-PCR | 5/6 (83%) | - | (Jiehao et al., 2020) |
| rRT-PCR | 1/1 (100%) | - | (Tang et al., 2020) |
Fig. 4.
Fluorescent probe-based RT-qPCR.
3.1.1. Developing a novel method
A passive sampling system may be a viable and affordable alternative to auto sampling due to the fact that the materials are deployed directly into wastewater to sorb viruses over time. The Moore swab (i.e. pieces of gauze tied to a fishing line) was used by Liu et al. (2020) to monitor SARS-CoV-2. Despite the fact that the swabs were deployed over a period of 24–72 hours, no evidence of accumulation of SARS-CoV-2 over time was observed. Three passive samplers (i.e., sorbents) were tested in a study conducted by Schang et al. (2020) for monitoring the presence of SARS-CoV-2 in sewage at larger scales (lot, suburb, and city). These materials were continuously exposed to flowing wastewater via a perforated device, resembling a torpedo, allowing for continuous use. SARS-CoV-2 samplers showed greater sensitivity than traditional samplers, especially when the concentrations were <1 copy/mL. In addition, Hayes et al. (2021) tested four different materials, including cotton gauze, cheesecloth, cellulose sponges, and electronegative membrane filters. Cheesecloth and membrane filters performed better (for accumulating SARS-CoV-2). There was no assessment, however, of the optimal deployment time (based on the concentration of SARS-CoV-2 in the wastewater).
4. Removing SARS-CoV-2 from wastewater
Firstly, the virus must be prevented from spreading before reclaimed water can be used. Water must be cleaned from coronaviruses in three steps before it can be used for recycling or reused. In the first phase of wastewater treatment, physical methods like screening and grit chambers remove suspended solids. The process of treatment is primarily conducted through biological treatment, with the remainder utilizing physicochemical treatment, which reduces turbidity, heavy metals, organics, and pathogens such as coronavirus (Gerba et al. 2019; Metcalf et al., 1991; Teymoorian et al., 2021). There is still a need for more extraordinary investigation into the effectiveness of these technologies for SARS-CoV-2. In addition, frequent evaluation of their performance in natural sewage treatment must evaluate the numerous aspects that impact viral survival and environmental effects in order to decide which methods are most efficient. Furthermore, (Zahmatkesh et al., 2022) prevented the disease from reaching the sewage system by reducing the trace amounts of SARS-CoV-2 in the stool with the use of hyssop. Finally, like with other coronaviruses, several variables, including pH, temperature, sunshine, and solids, significantly impact SARS-CoV-2.
4.1. Effect of physical processes on SARS-CoV-2
Separation by physical means is one approach to removing suspended volatile and fixed solids from sewage. After virus adsorption on large suspended solids, gravitational sedimentation is the dominant method of virus removal during the treatment phase. Despite this, gravity sedimentation does not eliminate viruses from wastewater. Coronavirus RNA is eradicated from secondary and tertiary wastewater treatment, and advanced wastewater is promptly utilized for agricultural and municipal purposes (Randazzo et al., 2020).
4.2. The impact of biological processes on SARS-CoV-2
There are many biological processes that are used in wastewater treatment plants to treat wastewater, including membrane bioreactors, activated sludge, and extended aeration (Randazzo et al., 2020). Studies conducted in the past found that secondary treatment significantly reduced the incidence of gastrointestinal viruses compared to the initial treatment method. According to another study, the primary treatment stage contains higher levels of organics that protect viruses, which increases the resistance and survival of coronaviruses. (Quilliam et al., 2020).
4.3. SARS-CoV-2 influence on biological process activated sludge and membrane
The uptake of viral particles effectively disinfects activated sludge-treated wastewater by organic biomass (Zahmatkesh et al., 2022; Zahmatkesh et al. 2020) and their sedimentation in the secondary clarifier (Foladori et al., 2021).
In secondary wastewater treatment, membrane bioreactors effectively remove viral particles. Filtration by membranes and growth by suspended particles comprise these processes. Compared to traditional processes, the membrane process is less expensive and more environmentally friendly, since very few chemicals are used, minimal technology is required, and it is simple to run (Obotey Ezugbe et al. 2020; Tetteh et al., 2020). Research demonstrates that this approach has significant restrictions due to its considerable power consumption, ranging from 0.45 to 0.65 kWh·m−3 for the most outstanding results (Obotey Ezugbe et al. 2020). The treatment of activated sludge (Abu Ali et al., 2021) is achieved in membrane bioreactor processes with longer solids retention times, which results in distinct treatment results. Membranes are more complex and have more significant operational challenges than activated sludge (Fortunato et al., 2019). As a result of the membrane bioreactor method, log reduction values of 6.8, 6.3, and 4.8 were achieved, respectively, for enteroviruses, adenoviruses, and noroviruses.
4.4. An assessment of the impact of the advanced process on SARS-CoV-2
In the tertiary phase, various treatment procedures are performed, including coagulation, filtration, ultraviolet light (UV), chlorine, and ozonation (Teymoorian et al., 2021). Virus inactivation and virus eradication in wastewater have been achieved with a variety of nanomaterials, including titanium dioxide, zero-valent iron, and carbon nanotubes (CNTs). As well as a genome, COVID-19 includes a protein capsule with an envelope or without it. A primary aim of viral sterilization methods (for example, UV, chlorination, and ozonization) was to impose environmental stress on one of these sections. Viral envelopes can be disrupted more easily. Therefore, non-enveloping viruses exhibit excellent resistance to inactivation and are less sensitive to adverse conditions (Fitzgibbon et al. 2008). A recent study conducted by Nasseri and colleagues has demonstrated that SARS-CoV 2 can be detected in 5 out of 6 water outlet samples when UV disinfection is used, and it can be detected in 1 out of 4 water outlet samples when chlorine disinfection is used. In this laboratory, only chlorine disinfection samples remained positive in the study (Nasseri et al., 2021). Thus, UV disinfection has been shown to be more effective than chlorine, and therefore WWTP operators should aim to reach a free residual chlorine concentration of 0.5 mg/L at all times.
5. Improves the efficiency of wastewater treatment by reducing the spread of SARS-CoV-2
The maximum temperature, light exposure, organic matter, and hostile bacteria are necessary for coronaviruses to live in water (Naddeo et al. 2020). Even though solar or UV light and temperatures above 23°C cause the virus to be inactivated, organic matter provides an adsorbent surface, increasing the virus's survival. Thus, it is vital to follow safe work practices and wear protective gear when working in the vicinity of untreated wastewater (Heilingloh et al., 2020; McGuigan et al., 2012).
5.1. Effects of solar, UV, and chlorination on SARS-CoV-2
UV disinfection machines are employed in public venues such as hospitals, airports, and shopping malls because they can disinfect regularly touched surfaces and streams of circulating air. Even so, UV treatment for water treatment offers a few advantages: it ensures clean disinfection, removes even some contaminants that can be treated relatively ineffectively, and is effective against most waterborne pathogens. The phosphodiester bonds, which form links between molecules and the viral genome, are reduced due to UV light exposure, depriving the infectious particles of their ability to replicate. Likewise, UV light is mainly efficient in disinfecting biologically polluted water. Other methods, such as chlorination, produce harmful by-products, but this procedure prevents microorganisms from growing in any medium. Depending upon their wavelength, UV photons consist of three types: UVA (315-400 nm), UVB (280-315 nm), and UVC (100–280 nm). (UVA photons have limited energy, while UVC photons may destroy the DNA of pathogens). In addition to UVA photons having minimal energy, UVC photons may also destroy pathogen DNA. By interacting with UVA and UVB wavelengths on Earth, UVC wavelengths, and pathogens, including viruses, may be damaged directly or indirectly via endogenous or exogenous components.
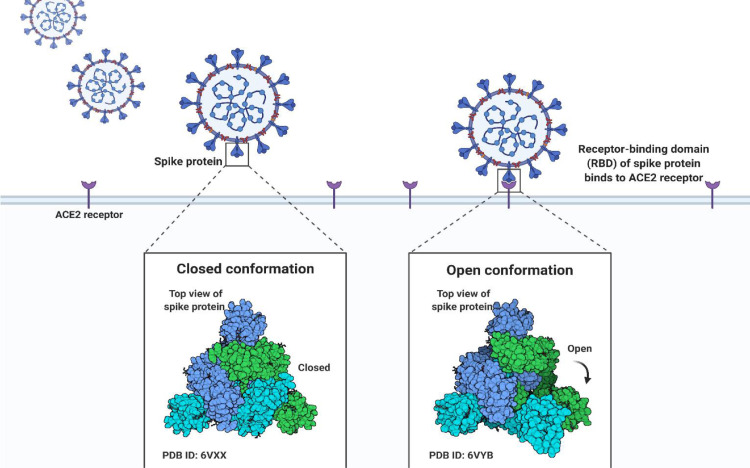
On the other hand, they are absorbed by the ozone layer and hence not visible on Earth. In solar water disinfection (SODIS), the amount of UV, heat, and duration are vital factors. In spite of this, the degree of exposure to sunlight (the amount of UV exposure) significantly influences viral disinfection more than time or temperature because prolonged exposure to heat only affect capsid proteins and have limited genome structure. Temperatures above 40°C may cause thermal-optical synergy, preventing each photon from inactivating more viruses. Consequently, the secondary treatment maintains residual protection, UV, and redundant protection against microbes ( Fig. 5 ) (Parsa, 2021; Parsa et al., 2021).
Fig. 5.
Analyzing the conformation of SARS-CoV-2 spike protein.
The UVA approach has been found in multiple investigations to be unsuccessful at eliminating SARS-CoV-1 (Darnell et al., 2004), whereas UVC eradicated the virus (Darnell et al., 2004; Parsa et al., 2021). Other investigations; however, failed to inactivate SARS-CoV-1 with UVC, the findings are ambiguous (Kariwa et al., 2006). Studies have been performed to determine whether UV irradiation may act as a preventative against the transmission of SARS-CoV-2 to individuals, explicitly employing UVC. By utilizing UVC wavelengths, it was possible to effectively remove surfaces infected with SARS-CoV-2 (Hadi et al., 2020; Heßling et al., 2020; Raeiszadeh et al. 2020). On diverse surfaces, UVC (222 nm) reduced SARS-CoV-2 concentrations by as much as 99.7%, based on Kitagawa et al. 's study (Kitagawa et al., 2021). At the same time, Heilingloh reported 1048 mg/cm2 as the best UVC dose to inactivate SARS-CoV-2 based on its studies (Heilingloh et al., 2020). Various types of UV seem to affect SARS-CoV-2 solutions differently. According to tests, UVA has a modest impact on deactivating SARS-CoV-2, but UVC is fully absorbed by the atmosphere and UVB only reaches the ground at a rate of 5% (Heilingloh et al., 2020). The UVC range of 260 to 265 nanometers is the most effective and optimum wavelength range for viral nucleic acids (Kowalski, 2010). When the ultraviolet wavelength is lower than 320 nm, the UVA wavelength is considered actinic, and at wavelengths above 320 nm, the virus nucleic acid is able to absorb fewer wavelengths, therefore this wavelength does not seem germicidal (Kowalski, 2010). Increasing wavelength decreases germicidal action, which reverses the relationship between germicidal action and wavelength (Pozo-Antonio et al. 2018).
Thermal energy is vital in solar disinfection, although its importance changes depending on the season. Due to the fact that thermal stress affects enzyme structure and activity above the optimum temperature for microorganisms, thermal stress has significant implications for health (Al-Gheethi et al., 2019). Most fecal bacteria, for example, can survive between 20 and 45°C (Marugán et al., 2020). Bacteria's ability to inactivate increases as temperatures rise above 45°C., whereas temperatures below 45°C do not significantly affect this process (McGuigan et al., 1998; Vivar et al., 2017). This means that SODIS is only capable of removing microorganisms in winter to a very limited extent. There is a direct relationship between volume, turbidity, and the environment with regard to thermal effects. Inhibition of DNA repair mechanisms is facilitated by thermal stress (McGuigan et al., 2012; Theitler et al., 2012). This leads to an increase in cell wall permeability (Theitler et al., 2012), leading to a reduction in enzyme activity and protein denaturation and cell death (Al-Gheethi et al., 2019; Marugán et al., 2020). As research has demonstrated, thermal and optical inactivation enhance the effectiveness of SODIS at temperatures more extraordinary than 45°C. However, it can only be beneficial in certain situations and under certain conditions (Castro-Alférez et al., 2017; McGuigan et al., 2012; Theitler et al., 2012; Vivar et al., 2017). Using the SODIS approach, the researchers assessed the temperature of the water to examine the influence of environmental factors on E. coli elimination. During summer, the water temperature fluctuates between 40 and 50 degrees Celsius . (Sichel et al., 2007). An examination of the effects of temperature, turbidity, and optical irradiation on the SODIS method has been conducted. According to the results of the research, pathogens must be completely inactivated at a temperature of 55°C (McGuigan et al., 1998). Despite this, experiments carried out in the field indicated that the surface temperature of the water reached only 45°C (Gómez-Couso et al., 2009). hus, using TiO2 photocatalysts in SODIS with and without photocatalysts produced water temperatures of 39°C and 38.6°C, respectively, while using a TiO2 photocatalyst produced water temperatures of 32.6°C and 36.6°C (Rincón et al. 2004). Furthermore, during colder seasons and warmer seasons, the actual water temperature is 15–21°C and 25–30°C, respectively (Sichel et al., 2007).
The process of disinfection produces free chlorine from several sources, including chlorine elements, chloramines, sodium hypochlorite, chlorine dioxide, calcium hypochlorite, and chloroisocyanurates. A highly successful approach to eliminating viral particles is using hypochlorous acid (HOCl) and hypochlorite ions (ClO−). Hypochlorite, a robust oxidizer, operates as an effective oxidizer of organic pollutants, whereas undissociated hypochlorous acid functions primarily as a microbicide. Based on a new investigation, the SARS-CoV was wholly inactivated after 30 minutes when the free residual chlorine amount was more than 0.4 mg/L and a free residual chlorine amount of about 2.19 mg/L (Wang et al., 2005). A recent study demonstrated that 1:99 diluted household bleach could inactivate the SARS-CoV-2 within five minutes when tested in vitro on SARS-CoV-2.
It is ammonia that is one of the most critical elements impacting effective chlorination, as chlorine helps take care of co-pollutants and pH. When chlorine interacts with ammonia, it generates a chlorine compound (chloramine), which is less effective at killing virus particles. Therefore, it is vital to prevent the absorption of Cl by any of the numerous substrates such as organic matter, ferrous ions, ammonia, hydrogen sulfide, and nitrites. Usually, organic elements mitigate the harmful effects of chlorinated chemicals, which represent short-term dangers to plants and soils (Ding et al., 2022).
A sampling of early studies revealed the presence of coronavirus RNA in the second and tertiary wastewater treatments. However, following the tertiary treatment of disinfectant treatment with NaClO, 100% of the samples were negative. However, the UV treatment was in addition to the disinfection procedure. The findings of Zhang et al. It was shown that sodium hypochlorite was capable of disinfecting SARS-CoV-2 viruses. This strategy, however, resulted in severe environmental pollution (Zhang et al., 2020).
6. Conclusion
During the outbreak of SAR-CoV-2, various research studies were conducted investigating the transmission of coronaviruses via polluted water or treated wastewater. Each investigation added to our knowledge of the virus's propagation in connection to water. During a COVID-19 pandemic, wastewater management is difficult and complex, and WBE and UV are critical approaches for treating sewage to prevent the dissemination of the virus.
The WBE may be applied as an additional tool to track COVID-19 instances among patients with SARS-CoV-2 and provide early detection of outbreaks. SARS-Co-2 is among numerous emerging severe infectious viruses in sewage that bring novel obstacles and potential when employing WBE for its monitoring. Finally, a viable WBE technique depends on a representative sample, virus quantities in wastewater, the averaging of the population, and ethical norms.
Based on the identification of SARS-CoV-2 in feces and the widespread transmission of the virus as demonstrated by virological tests, a WHO recommended that the WBE strategy be developed to minimize and prevent the transmission of COVID-19. Based on WBE, the original concentration of any stable substance emitted by the serviced community can be back calculated if humans excrete it in wastewater. The same approach can also be applied to examine the circulation of pathogens in sewers, which are typically excreted by infected individuals in their feces or urine. Whenever clinical assessment resources are scarce or information resources are lacking, this approach is advantageous.
Funding information
This work did not receive any financial support.
References
- Abu Ali H., Yaniv K., Bar-Zeev E., Chaudhury S., Shagan M., Lakkakula S., Ronen Z., Kushmaro A., Nir O. Tracking SARS-CoV-2 RNA through the wastewater treatment process. ACS ES&T Water. 2021;1(5):1161–1167. doi: 10.1021/acsestwater.0c00216. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Gheethi A.A.S., Noman E.A., Mohamed R.M.S.R., Talip B.A., Kassim A.H.M., Ismail N. Springer; 2019. Disinfection Technologies for Household Greywater. Management of Greywater in Developing Countries; pp. 185–203. [Google Scholar]
- Asghar H., Diop O.M., Weldegebriel G., Malik F., Shetty S., El Bassioni L., Akande A.O., Al Maamoun E., Zaidi S., Adeniji A.J. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014;210(suppl_1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilal M., Nazir M.S., Parra-Saldivar R., Iqbal H. NCOV/COVID-19—approaches to viral vaccine development and preventive measures. J. Pure Appl. Microbiol. 2020;14(1):25–29. [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.-M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.-L. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Eurosurveillance. 2018;23(7):17–00237. doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno Ferraro G., Mancini P., Veneri C., Iaconelli M., Suffredini E., Brandtner D., La Rosa G. Evidence of Saffold virus circulation in Italy provided through environmental surveillance. Lett. Appl. Microbiol. 2020;70(2):102–108. doi: 10.1111/lam.13249. [DOI] [PubMed] [Google Scholar]
- Bugembe D.L., Phan M.V., Abias A.G., Ayei J., Deng L.L., Lako R.L.L., Rumunu J., Kaleebu P., Wamala J.F., Hm J.J. SARS-CoV-2 variants, South Sudan, January–March 2021. Emerg. Infect. Dis. 2021;27(12):3133. doi: 10.3201/eid2712.211488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B., Rodriguez-Manzano J., Hundesa A., Suñen E., Calvo M., Bofill-Mas S., Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods. 2013;187(2):215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Carducci A., Verani M., Battistini R., Pizzi F., Rovini E., Andreoli E., Casini B. Epidemiological surveillance of human enteric viruses by monitoring of different environmental matrices. Water Sci. Technol. 2006;54(3):239–244. doi: 10.2166/wst.2006.475. [DOI] [PubMed] [Google Scholar]
- Cashdollar J., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115(1):1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- Castro-Alférez M., Polo-López M.I., Marugán J., Fernandez-Ibanez P. Mechanistic model of the Escherichia coli inactivation by solar disinfection based on the photo-generation of internal ROS and the photo-inactivation of enzymes: CAT and SOD. Chem. Eng. J. 2017;318:214–223. [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chen Y., Feng W., Ye K., Guo L., Xia H., Guan Y., Chai L., Shi W., Zhai C., Wang J. Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell. Infect. Microbiol. 2021;11:168. doi: 10.3389/fcimb.2021.541092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F., Chan P.P., Lung K., Tso E., Liu R., Ng Y., Chu M.Y., Chung T.W., Tam A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus, N. (2019). Real-time rRT-PCR panel primers and probes, US Centers for Disease Control and Prevention, https://www.cdc.gov
- COVID C., Team R. SARS-CoV-2 B. 1.1. 529 (Omicron) variant—United States, December 1–8, 2021. Morb. Mortal. Wkly Rep. 2021;70(50):1731. doi: 10.15585/mmwr.mm7050e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A., Kantor R.S., Olm M.R., Whitney O.N., Al-Shayeb B., Lou Y.C., Flamholz A., Kennedy L.C., Greenwald H., Hinkle A. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. MBio. 2021;12(1):E02703–E02720. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods. 2004;121(1):85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Pu Y., Tang M., Zhang T. Environmental and health effects of graphene-family nanomaterials: potential release pathways, transformation, environmental fate and health risks. Nano Today. 2022;42 [Google Scholar]
- Dubey A., Choudhary S., Kumar P., Tomar S. Emerging SARS-CoV-2 variants: genetic variability and clinical implications. Curr. Microbiol. 2022;79(1):1–18. doi: 10.1007/s00284-021-02724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré V.M., Peiffer-Smadja N., Visseaux B., Descamps D., Ghosn J., Charpentier C. Omicron SARS-CoV-2 variant: what we know and what we don't. Anaesthesia, Crit. Care Pain Med. 2022;41(1) doi: 10.1016/j.accpm.2021.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J., Sagripanti J.L. Analysis of the survival of Venezuelan equine encephalomyelitis virus and possible viral simulants in liquid suspensions. J. Appl. Microbiol. 2008;105(5):1477–1483. doi: 10.1111/j.1365-2672.2008.03919.x. [DOI] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Cadonna M., Manara S. Coronaviruses and SARS-CoV-2 in sewerage and their removal: step by step in wastewater treatment plants. Environ. Res. 2021 doi: 10.1016/j.envres.2021.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.-T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69(2):357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenele, R. S., Kraberger, S., Hadfield, J., Driver, E. M., Bowes, D., Holland, L. A., Faleye, T. O., Adhikari, S., Kumar, R., & Inchausti, R. (2021). “High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants.” MedRxiv. [DOI] [PMC free article] [PubMed]
- Fortunato L., Li M., Cheng T., Rehman Z.U., Heidrich W., Leiknes T. Cake layer characterization in activated sludge membrane bioreactors: real-time analysis. J. Membr. Sci. 2019;578:163–171. [Google Scholar]
- Fumian T.M., Leite J.P.G., Castello A.A., Gaggero A., de Caillou M.S.L., Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170(1-2):42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Gerba C.P., Pepper I.L. Environmental and Pollution Science. Elsevier; 2019. Municipal wastewater treatment; pp. 393–418. [Google Scholar]
- Gómez-Couso H., Fontán-Saínz M., Sichel C., Fernández-Ibáñez P., Ares-Mazás E. Efficacy of the solar water disinfection method in turbid waters experimentally contaminated with Cryptosporidium parvum oocysts under real field conditions. Tropical Medicine & International Health. 2009;14(6):620–627. doi: 10.1111/j.1365-3156.2009.02281.x. [DOI] [PubMed] [Google Scholar]
- Graber T.E., Mercier É., Bhatnagar K., Fuzzen M., D'Aoust P.M., Hoang H.-D., Tian X., Towhid S.T., Plaza-Diaz J., Eid W. Near real-time determination of B. 1.1. 7 in proportion to total SARS-CoV-2 viral load in wastewater using an allele-specific primer extension PCR strategy. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Deng X., Lee M., Sucu Y.D., Arevalo S., Stryke D., Federman S., Gopez A., Reyes K., Zorn K. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 2021;27(1):115–124. doi: 10.1038/s41591-020-1105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi J., Dunowska M., Wu S., Brightwell G. Control measures for sars-cov-2: a review on light-based inactivation of single-stranded rna viruses. Pathogens. 2020;9(9):737. doi: 10.3390/pathogens9090737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 2009;32(3):297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilingloh C.S., Aufderhorst U.W., Schipper L., Dittmer U., Witzke O., Yang D., Zheng X., Sutter K., Trilling M., Alt M. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control. 2020;48(10):1273–1275. doi: 10.1016/j.ajic.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heßling M., Hönes K., Vatter P., Lingenfelder C. Ultraviolet irradiation doses for coronavirus inactivation–review and analysis of coronavirus photoinactivation studies. GMS Hyg. Infect. Control. 2020;15 doi: 10.3205/dgkh000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V.R., Polaczyk A.L., Hahn D., Narayanan J., Cromeans T.L., Roberts J.M., Amburgey J.E. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 2005;71(11):6878–6884. doi: 10.1128/AEM.71.11.6878-6884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrudey S.E., Conant B. The devil is in the details: emerging insights on the relevance of wastewater surveillance for SARS-CoV-2 to public health. J. Water Health. 2021 doi: 10.2166/wh.2021.186. [DOI] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4(2):41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Jiehao C., Jin X., Daojiong L., Zhi Y., Lei X., Zhenghai Q., Yuehua Z., Hua Z., Ran J., Pengcheng L. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;71(6):1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.A., Vasconcelos P.F., Staples J.E. The whole iceberg: estimating the incidence of yellow fever virus infection from the number of severe cases. Trans. R. Soc. Trop. Med. Hyg. 2014;108(8):482–487. doi: 10.1093/trstmh/tru092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariwa H., Fujii N., Takashima I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology. 2006;212(suppl. 1):119–123. doi: 10.1159/000089211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Chung H.-S., Noh S.G., Lee B., Chung H.Y., Choi J.-G. Geraniin inhibits the entry of SARS-CoV-2 by blocking the interaction between spike protein RBD and human ACE2 receptor. Int. J. Mol. Sci. 2021;22(16):8604. doi: 10.3390/ijms22168604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H., Nomura T., Nazmul T., Omori K., Shigemoto N., Sakaguchi T., Ohge H. Effectiveness of 222-nm ultraviolet light on disinfecting SARS-CoV-2 surface contamination. Am. J. Infect. Control. 2021;49(3):299–301. doi: 10.1016/j.ajic.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Rachmadi A.T., Haramoto E., Gerba C.P. Quantification and genetic analysis of salivirus/klassevirus in wastewater in Arizona, USA. Food Environ. Virol. 2014;6(3):213–216. doi: 10.1007/s12560-014-9148-2. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Pepper I.L., Gerba C.P. Occurrence and genetic diversity of human cosavirus in influent and effluent of wastewater treatment plants in Arizona, United States. Arch. Virol. 2015;160(7):1775–1779. doi: 10.1007/s00705-015-2435-x. [DOI] [PubMed] [Google Scholar]
- Kowalski W. Springer science & business media; 2010. Ultraviolet Germicidal Irradiation Handbook: UVGI for Air and Surface Disinfection. [Google Scholar]
- La Rosa G., Della Libera S., Iaconelli M., Ciccaglione A.R., Bruni R., Taffon S., Equestre M., Alfonsi V., Rizzo C., Tosti M.E. Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013. BMC Infect. Dis. 2014;14(1):1–11. doi: 10.1186/1471-2334-14-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Muscillo M. Viruses in Food and Water. Elsevier; 2013. Molecular detection of viruses in water and sewage; pp. 97–125. [Google Scholar]
- Lai C.-C., Liu Y.H., Wang C.-Y., Wang Y.-H., Hsueh S.-C., Yen M.-Y., Ko W.-C., Hsueh P.-R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 2020;53(3):404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. L., Gu, X., Armas, F., Wu, F., Chandra, F., Chen, H., Xiao, A., Leifels, M., Chua, D. F., & Kwok, G. W. (2021). "Quantitative detection of SARS-CoV-2 Omicron variant in wastewater through allele-specific RT-qPCR." MedRxiv.
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54(8):1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu Y., Yu Y., Zhang X., Li B., Wu J., Li J., Wu Y., Xia X., Tang H. Positive result of Sars-Cov-2 in faeces and sputum from discharged patients with COVID-19 in Yiwu, China. J. Med. Virol. 2020;92(10):1938–1947. doi: 10.1002/jmv.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., Tian Y., Sin N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16(10):1698. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugán J., Giannakis S., McGuigan K.G., Polo-López I. Solar disinfection as a water treatment technology. Clean Water Sanitation. 2020:1–16. [Google Scholar]
- McGuigan K., Joyce T.M., Conroy R.M., Gillespie J., Elmore-Meegan M. Solar disinfection of drinking water contained in transparent plastic bottles: characterizing the bacterial inactivation process. J. Appl. Microbiol. 1998;84(6):1138–1148. doi: 10.1046/j.1365-2672.1998.00455.x. [DOI] [PubMed] [Google Scholar]
- McGuigan K.G., Conroy R.M., Mosler H.-J., du Preez M., Ubomba-Jaswa E., Fernandez-Ibanez P. Solar water disinfection (SODIS): a review from bench-top to roof-top. J. Hazard. Mater. 2012;235:29–46. doi: 10.1016/j.jhazmat.2012.07.053. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Mehra R., Kepp K.P. Structure and mutations of SARS-CoV-2 spike protein: a focused overview. ACS Infect. Dis. 2021;8(1):29–58. doi: 10.1021/acsinfecdis.1c00433. [DOI] [PubMed] [Google Scholar]
- Metcalf L., Eddy H.P., Tchobanoglous G. McGraw-Hill; New York: 1991. Wastewater Engineering: Treatment, Disposal, and Reuse. [Google Scholar]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109(2):388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25(10) doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020;54(9):5311. doi: 10.1021/acs.est.0c02172. -5311. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. 2020;6(5):1213–1216. [Google Scholar]
- Nagy A., Basiouni S., Parvin R., Hafez H.M., Shehata A.A. Evolutionary insights into the furin cleavage sites of SARS-CoV-2 variants from humans and animals. Arch. Virol. 2021;166(9):2541–2549. doi: 10.1007/s00705-021-05166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napit, R., Manandhar, P., Chaudhary, A., Shrestha, B., Poudel, A., Raut, R., Pradhan, S., Raut, S., Mathema, S., & Rajbhandari, R. (2021). "Rapid genomic surveillance of SARS-CoV-2 in a dense urban community using environmental (sewage) samples." MedRxiv. [DOI] [PMC free article] [PubMed]
- Nasseri S., Yavarian J., Baghani A.N., Azad T.M., Nejati A., Nabizadeh R., Hadi M., Jandaghi N.Z.S., Vakili B., Vaghefi S.K.A. The presence of SARS-CoV-2 in raw and treated wastewater in 3 cities of Iran: Tehran, Qom and Anzali during coronavirus disease 2019 (COVID-19) outbreak. J. Environ. Health Sci. Eng. 2021;19(1):573–584. doi: 10.1007/s40201-021-00629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung S.-m., Hayashi K., Kinoshita R., Yang Y., Yuan B., Akhmetzhanov A.R. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obotey Ezugbe E., Rathilal S. Membrane technologies in wastewater treatment: a review. Membranes. 2020;10(5):89. doi: 10.3390/membranes10050089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi T., Yokota S.-i., Ohkoshi Y., Ohuchi H., Yoshida Y., Kikuchi M., Yano K., Fujii N. Occurrence of norovirus infections unrelated to norovirus outbreaks in an asymptomatic food handler population. J. Clin. Microbiol. 2008;46(6):1985–1988. doi: 10.1128/JCM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. World Health Organization; 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases: Interim Guidance. 14 January 2020. [Google Scholar]
- Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3) doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa S.M. Reliability of thermal desalination (solar stills) for water/wastewater treatment in light of COVID-19 (novel coronavirus “SARS-CoV-2”) pandemic: what should consider? Desalination. 2021;512 doi: 10.1016/j.desal.2021.115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa S.M., Momeni S., Hemmat A., Afrand M. Effectiveness of solar water disinfection in the era of COVID-19 (SARS-CoV-2) pandemic for contaminated water/wastewater treatment considering UV effect and temperature. J. Water Process Eng. 2021;43 doi: 10.1016/j.jwpe.2021.102224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19–approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo-Antonio J.S., Sanmartín P. Exposure to artificial daylight or UV irradiation (A, B or C) prior to chemical cleaning: an effective combination for removing phototrophs from granite. Biofouling. 2018;34(8):851–869. doi: 10.1080/08927014.2018.1512103. [DOI] [PubMed] [Google Scholar]
- Prevost B., Lucas F., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Prüss A., Kay D., Fewtrell L., Bartram J. Estimating the burden of disease from water, sanitation, and hygiene at a global level. Environ. Health Perspect. 2002;110(5):537–542. doi: 10.1289/ehp.110-1240845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam, J. R., van Schalkwyk, C., Govender, N., von Gottberg, A., Cohen, C., Groome, M. J., Dushoff, J., Mlisana, K., & Moultrie, H. (2021). "Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa." MedRxiv. [DOI] [PMC free article] [PubMed]
- Qi R., Huang Y.-t., Liu J.-w., Sun Y., Sun X.-f., Han H.-J., Qin X.-R., Zhao M., Wang L.-j., Li W. Global prevalence of asymptomatic norovirus infection: a meta-analysis. EClinicalMedicine. 2018;2:50–58. doi: 10.1016/j.eclinm.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam R.S., Weidmann M., Moresco V., Purshouse H., O'Hara Z., Oliver D.M. COVID-19: the environmental implications of shedding SARS-CoV-2 in human faeces. Environ. Int. 2020;140 doi: 10.1016/j.envint.2020.105790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeiszadeh M., Adeli B. A critical review on ultraviolet disinfection systems against COVID-19 outbreak: applicability, validation, and safety considerations. Acs Photonics. 2020;7(11):2941–2951. doi: 10.1021/acsphotonics.0c01245. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón A.-G., Pulgarin C. Field solar E. coli inactivation in the absence and presence of TiO2: is UV solar dose an appropriate parameter for standardization of water solar disinfection? Sol. Energy. 2004;77(5):635–648. [Google Scholar]
- Rodriguez-Lazaro D., Cook N., Ruggeri F.M., Sellwood J., Nasser A., Nascimento M.S.J., D'Agostino M., Santos R., Saiz J.C., Rzeżutka A. Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 2012;36(4):786–814. doi: 10.1111/j.1574-6976.2011.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen-Kopra C., Paananen A., Blomqvist S., Klemola P., Simonen M.-L., Lappalainen M., Vuorinen T., Kuusi M., Lemey P., Roivainen M. A large Finnish echovirus 30 outbreak was preceded by silent circulation of the same genotype. Virus Genes. 2011;42(1):28–36. doi: 10.1007/s11262-010-0536-x. [DOI] [PubMed] [Google Scholar]
- Sedmak G., Bina D., MacDonald J. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 2003;69(12):7181–7187. doi: 10.1128/AEM.69.12.7181-7187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Japanese J. Infect. Dis.: JJID. 2020 doi: 10.7883/yoken.JJID.2020.061. 2020.2061. [DOI] [PubMed] [Google Scholar]
- Sichel C., Blanco J., Malato S., Fernandez-Ibanez P. Effects of experimental conditions on E. coli survival during solar photocatalytic water disinfection. J. Photochem. Photobiol. A. 2007;189(2-3):239–246. [Google Scholar]
- Sichel C., Tello J., De Cara M., Fernández-Ibáñez P. Effect of UV solar intensity and dose on the photocatalytic disinfection of bacteria and fungi. Catal. Today. 2007;129(1-2):152–160. [Google Scholar]
- Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.M., Taha T.Y., Tabata T., Chen I.P., Ciling A., Khalid M.M., Sreekumar B., Chen P.-Y., Hayashi J.M., Soczek K.M. Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles. Science. 2021;374(6575):1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.-d., Wang H.-l., Dai Y.-x., Li K.-f., Liu J.-n., Wu W.-j., Yuan C., Yu M.-l., Li P. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26(6):1337. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh E.K., Obotey Ezugbe E., Rathilal S., Asante-Sackey D. Removal of COD and SO42− from oil refinery wastewater using a photo-catalytic system—comparing tio2 and zeolite efficiencies. Water. 2020;12(1):214. [Google Scholar]
- Teymoorian T., Teymourian T., Kowsari E., Ramakrishna S. Direct and indirect effects of SARS-CoV-2 on wastewater treatment. J. Water Process Eng. 2021;42 doi: 10.1016/j.jwpe.2021.102193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theitler D.J., Nasser A., Gerchman Y., Kribus A., Mamane H. Synergistic effect of heat and solar UV on DNA damage and water disinfection of E. coli and bacteriophage MS2. J. Water Health. 2012;10(4):605–618. doi: 10.2166/wh.2012.072. [DOI] [PubMed] [Google Scholar]
- Thompson R.N. Novel coronavirus outbreak in Wuhan, China, 2020: intense surveillance is vital for preventing sustained transmission in new locations. J. Clin. Med. 2020;9(2):498. doi: 10.3390/jcm9020498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongprachum A., Fujimoto T., Takanashi S., Saito H., Okitsu S., Shimizu H., Khamrin P., Maneekarn N., Hayakawa S., Ushijima H. Detection of nineteen enteric viruses in raw sewage in Japan. Infect. Genet. Evol. 2018;63:17–23. doi: 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Trimpert J., Adler J.M., Eschke K., Abdelgawad A., Firsching T.C., Ebert N., Thao T.T.N., Gruber A.D., Thiel V., Osterrieder N. Live attenuated virus vaccine protects against SARS-CoV-2 variants of concern B. 1.1. 7 (Alpha) and B. 1.351 (Beta) Sci. Adv. 2021;7(49):Eabk0172. doi: 10.1126/sciadv.abk0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Poelvoorde L.A., Gand M., Fraiture M.-A., De Keersmaecker S.C., Verhaegen B., Van Hoorde K., Cay A.B., Balmelle N., Herman P., Roosens N. Strategy to develop and evaluate a multiplex RT-ddPCR in response to SARS-CoV-2 genomic evolution. Curr. Issues Mol. Biol. 2021;43(3):1937–1949. doi: 10.3390/cimb43030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers M.F., Cohen A.F., Van Gerven J.M., Groeneveld G.J. The impact of the global COVID-19 pandemic on the conduct of clinical trials: return to normalcy by considering the practical impact of a structured ethical analysis. Br. J. Clin. Pharmacol. 2021;87(3):837–844. doi: 10.1111/bcp.14480. [DOI] [PubMed] [Google Scholar]
- Vivar M., Pichel N., Fuentes M. Solar disinfection of natural river water with low microbiological content (10–103 CFU/100 ml) and evaluation of the thermal contribution to water purification. Sol. Energy. 2017;141:1–10. [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126(1-2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems. 2020;5(4):E00614–E00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer, S., Marechal, V., Mouchel, J.-M., & Moulin, L. (2020). "Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases." MedRxiv.
- Wyn-Jones A., Sellwood J. Enteric viruses in the aquatic environment. J. Appl. Microbiol. 2001;91(6):945–962. doi: 10.1046/j.1365-2672.2001.01470.x. [DOI] [PubMed] [Google Scholar]
- Xagoraraki I., O'Brien E. Women in Water Quality. Springer; 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [Google Scholar]
- Xia X. Domains and functions of spike protein in sars-cov-2 in the context of vaccine design. Viruses. 2021;13(1):109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Lewis Y., Kushmaro A. RT-qPCR assays for SARS-CoV-2 variants of concern in wastewater reveals compromised vaccination-induced immunity. Water Res. 2021;207 doi: 10.1016/j.watres.2021.117808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zahmatkesh S., Amesho K.T., Sillanpää M. A critical review on diverse technologies for advanced wastewater treatment during SARS-CoV-2 pandemic: what do we know? J. Hazard. Mater. Adv. 2022;7 doi: 10.1016/j.hazadv.2022.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahmatkesh S., Amesho K.T., Sillanpaa M., Wang C. Integration of renewable energy in wastewater treatment during COVID-19 pandemic: challenges, opportunities, and progressive research trends. Clean. Chem. Eng. 2022;3 [Google Scholar]
- Zahmatkesh S., Far S.S., Sillanpää M. RSM-D-optimal modeling approach for COD removal from low strength wastewater by microalgae, sludge, and activated carbon-case study mashhad. J. Hazard. Mater. Adv. 2022;7 [Google Scholar]
- Zahmatkesh S., Klemeš J.J., Bokhari A., Wang C., Sillanpaa M., Hasan M., Amesho K.T. Critical role of Hyssop plant in the possible transmission of SARS-CoV-2 in contaminated human Feces and its implications for the prevention of the virus spread in sewage. Chemosphere. 2022;305 doi: 10.1016/j.chemosphere.2022.135247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahmatkesh S., Klemeš J.J., Bokhari A., Wang C., Sillanpaa M., Amesho K. Reducing Chemical Oxygen Demand from Low Strength Wastewater: A Novel Application of Fuzzy Logic Based Simulation in MATLAB. Computers & Chemical Engineering. 2022 [Google Scholar]
- Zahmatkesh S., Pirouzi A. Effects of the microalgae, sludge and activated carbon on the wastewater treatment with low organics (weak wastewater) Int. J. Environ. Sci. Technol. 2020;17(5):2681–2688. [Google Scholar]
- Zahmatkesh S., Sillanpää M. Review of method and a new tool for decline and inactive SARS-CoV-2 in wastewater treatment. Clean. Chem. Eng. 2022;3 [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang S., Xue Y. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020;92(6):680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. e882. [DOI] [PMC free article] [PubMed] [Google Scholar]