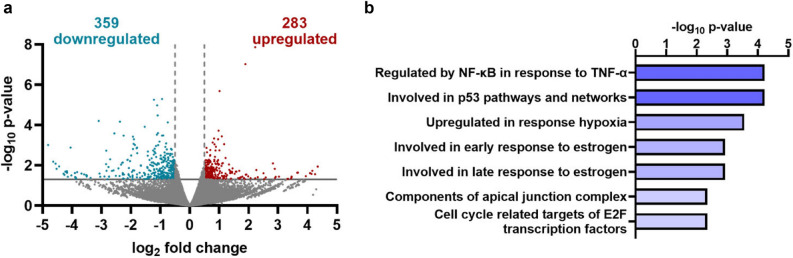
Figure 1.
Altered gene expression associated with ADAR3 expression in U87 cells. (a) Dots represent individual genes that are downregulated (blue), upregulated (red), or not significantly different (gray) in ADAR3-expressing U87 cells compared to control U87 cells in three biological replicates of RNA-seq data. Negative log10 p-values of differential expression (y-axis) are plotted against the average log2 fold change in expression (x-axis). Genes considered significantly differentially expressed exhibited log2 fold change of 0.5 (light gray broken vertical lines) and a p-value of 0.05 [log10 p-value of 1.3, light gray horizontal line]. (b) Significantly enriched pathways for the 641 differentially expressed genes are listed in descending order of the -log10 p-value provided by the Gene Set Enrichment Analysis computational method.