FIGURE 1.
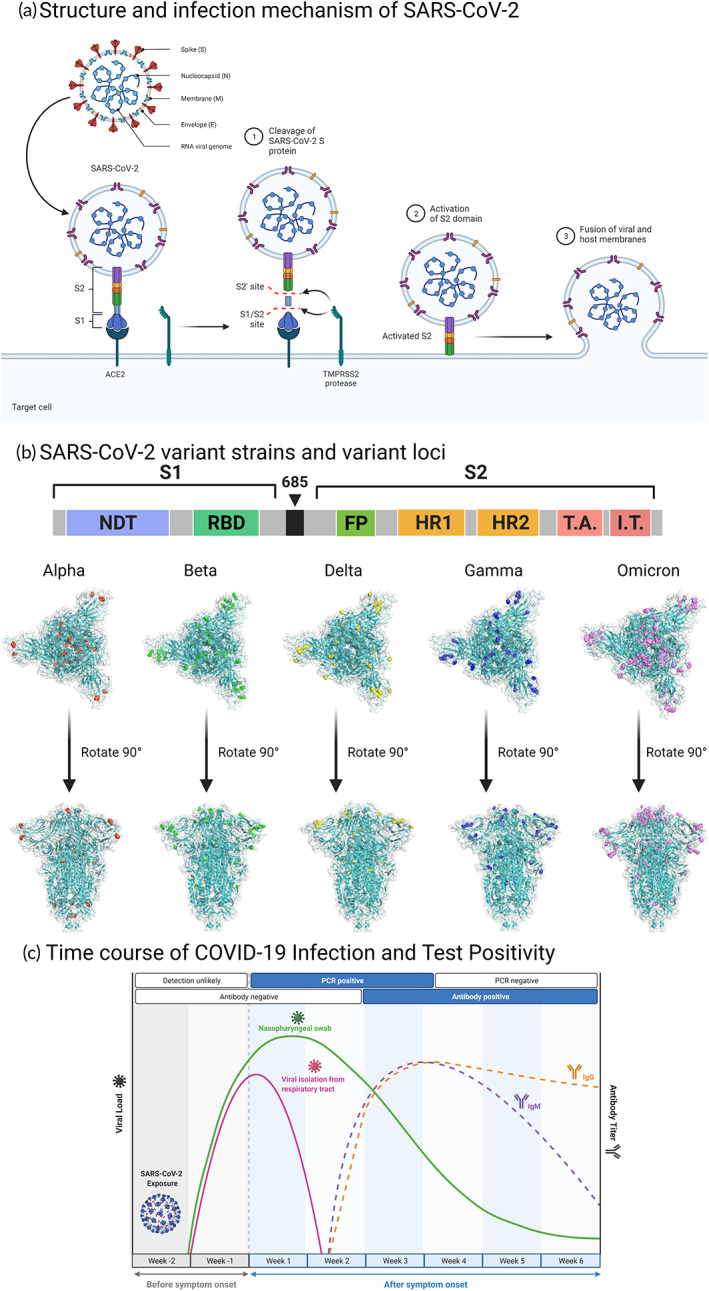
Biology and serology of SARS‐CoV‐2 infection (a) Structure and infection: SARS‐CoV‐2 is an RNA virus that consists of four structural proteins, the Spike (S) protein, Nucleocapsid (N) protein, Membrane (M) protein, and Envelope (e), together with many non‐structural proteins to maintain the biological traits of the virus. Step 1–3: S protein allows the virus to bind and enter human cells and consists of S1 and S2 subunits. S1 can bind the angiotensin‐converting enzyme 2 (ACE2) receptor. After S1 binds to ACE2, S protein is hydrolyzed by the action of TMPRSS2 protease. The activated S2 subunit can then further mediate the fusion of membranes between the host cell and the virus, allowing the virus to enter the host cell. (b) SARS‐CoV‐2 variants: S protein of the first Wuhan‐Hu‐1 strain consisted of 1273 amino acid residues, in which the S1 and S2 fragments are linked by amino acid bridges, S1 includes the N‐terminal domain (NTD) and receptor‐binding domain (RBD), and S2 includes the fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), and other structures. Since the start of the outbreak, many strongly infectious SARS‐CoV‐2 mutant strains have emerged, such as B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.1.529 (Omicron), among which mutations are particularly common in the S protein and have a substantial effect on the infectivity of the virus. (PDB ID:7DDD) (c) Immunity: Following viral infection in humans, specific antibody reactions often appear between days 5 and 15 after infection, with the IgM response lasting 3–6 weeks and the IgG response lasting several months.
