FIGURE 2.
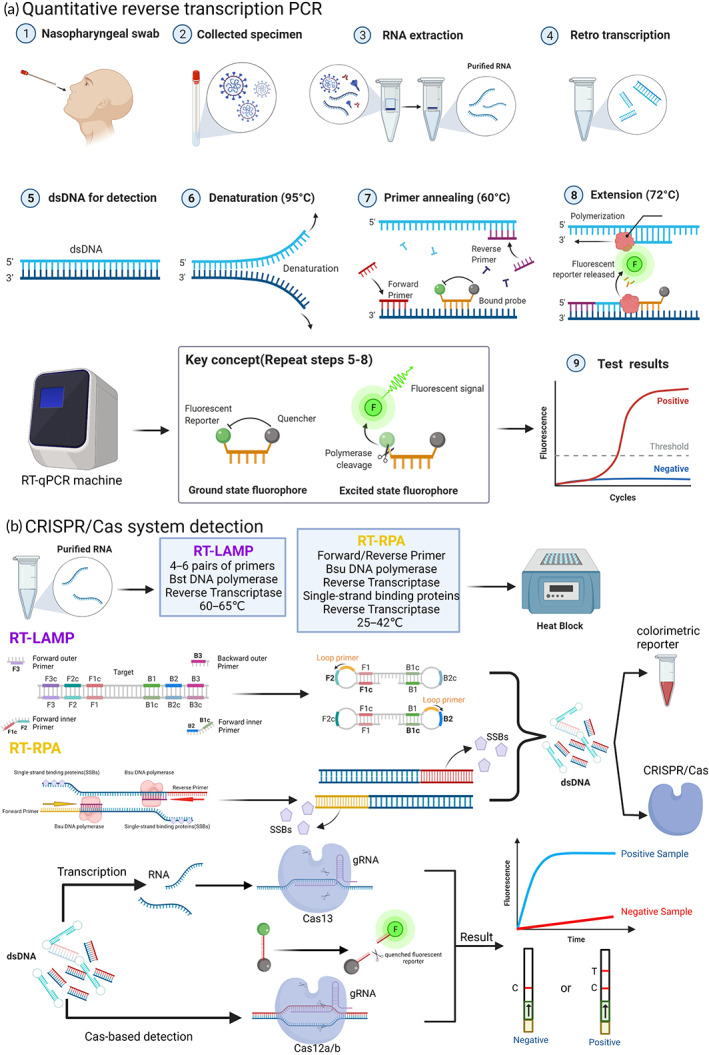
Nucleic acid‐based detection of SARS‐CoV‐2 (a) qRT‐PCR: Step 1–4: SARS‐CoV‐2 RNA in different collected samples, such as nasopharyngeal swabs, can be extracted and purified using an RNA extraction kit, and complementary DNA (cDNA) for amplification and detection can be obtained by reverse transcriptase; Step 5–9: template cDNA undergoes denaturation, primer annealing, and extension in the real‐time PCR instrument The fluorescence signal is released when the fluorescence molecule is no longer inhibited by the quenching molecule, and the instrument can convert the fluorescence signal in the cycle into the cycle threshold (CT) value, which can be expressed as the quantified viral load data, and the validity of SARS‐CoV‐2 infection is verified by comparison with negative controls and threshold lines. (b) CRISPR/Cas system: Based on reverse transcription recombinant polymerase amplification (RT‐RPA) and reverse transcription loop‐mediated isothermal amplification (RT‐LAMP), purified RNA can be amplified in an isothermal instrument, and the amplified product can be reported both by the chromogenic substances in the amplification system and by the CRISPR/Cas system for further specific cleavage of nucleic acids and determination of virus infection. The CRISPR‐associated Cas protein then binds to the guide RNA, forming a complex that can target cleavage of the viral nucleic acid sequence, and the result can be reported by the fluorescence quenching molecules in the reaction, by reporting the fluorescence signal, or by the side stream chromatography color development strip of the cleaved nucleic acid fragment.
