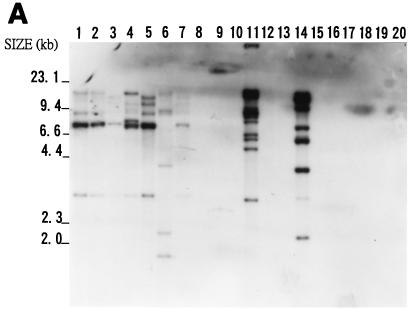
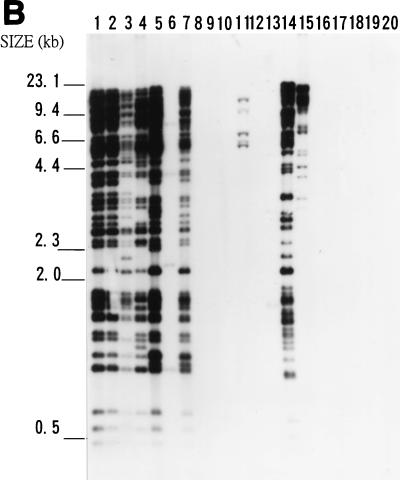
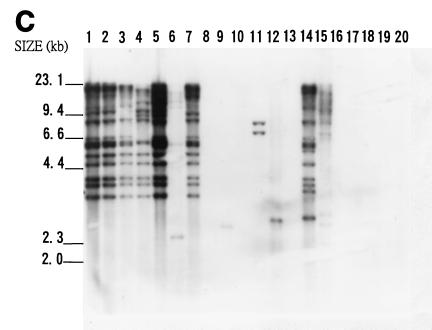
FIG. 5.
Southern blot hybridization analyses of total DNAs from 20 bacteria. Samples of about 3 μg of EcoRI-digested genomic DNAs were analyzed on an agarose gel, transferred to a nylon membrane, and hybridized with the IS476A probe (A), IS1478A probe (B), or IS1479A probe (C). DNAs from X. campestris pv. campestris 11A (lane 1), X. campestris pv. campestris 11 (lane 2), X. campestris pv. campestris 17 (lane 3), X. campestris pv. campestris 2 (lane 4), X. campestris pv. campestris 6 (lane 5), X. campestris pv. campestris 85 (lane 6), X. campestris pv. campestris 88 (lane 7), X. campestris pv. mangiferaeindicae 38 (lane 8), X. campestris pv. glycinea 69 (lane 9), X. campestris pv. dieffenbachiae 65 (lane 10), X. campestris pv. vesicatoria 64 (lane 11), X. campestris pv. phaseoli 73 (lane 12), X. campestris pv. citri (lane 13), X. campestris pv. begoniae (lane 14), X. campestris pv. oryzae 1 (lane 15), Agrobacterium tumefaciens LBA4404 (lane 16), Erwinia carotovora subsp. carotovora ZL4 (lane 17), Pseudomonas solanacearum RD4 (lane 18), Rhizobium leguminosarum 128C53 (lane 19), and E. coli DH5α (lane 20) were analyzed. The sizes of HindIII-digested λ DNA fragments are indicated to the left of the gels.