Figure 1.
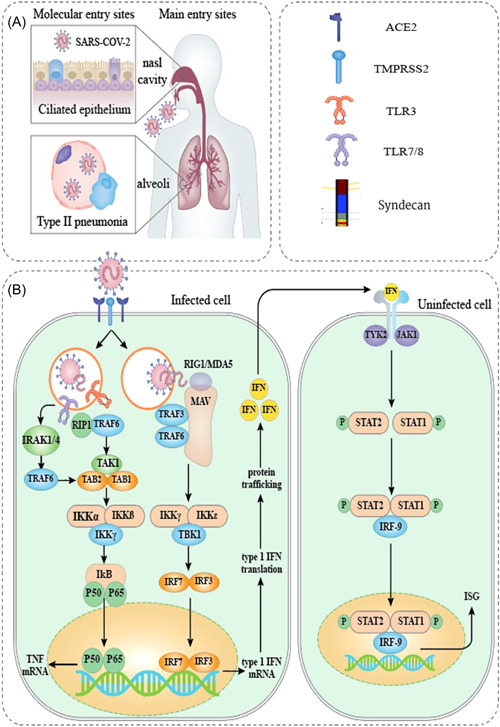
. The schematic representation of SARS‐CoV‐2 pathophysiology. (A) SARS‐CoV‐2 enters the body primarily through cells in the nasal cavity and the upper and lower respiratory tracts. (B) Several PRRs that identify foreign RNA, such as endosomal TLR3 and TLR7, and cytoplasmic RIG‐I and MDA5, are thought to be involved in recognizing SARS‐CoV‐2. Results from genetic research, functional and clinical findings, interaction modeling, and CRISPR screens are used to estimate downstream signaling occurrences. Direct communication among viral or host proteins and interplay among SARS‐CoV‐2‐derived proteins and cellular mechanisms as defined by interaction mapping derived information. ORF3b was found to be functionally active in the suppression of type I IFN, but no specific target was recognized. 12 CRISPR, clustered regularly interspaced short palindromic repeat; IFN, interferon; MDA5, melanoma differentiation‐associated protein 5; ORF, open reading frame; PRR, pattern recognition receptor; TLR, Toll‐like receptor; RIG‐I, retinoic acid‐inducible gene I; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
