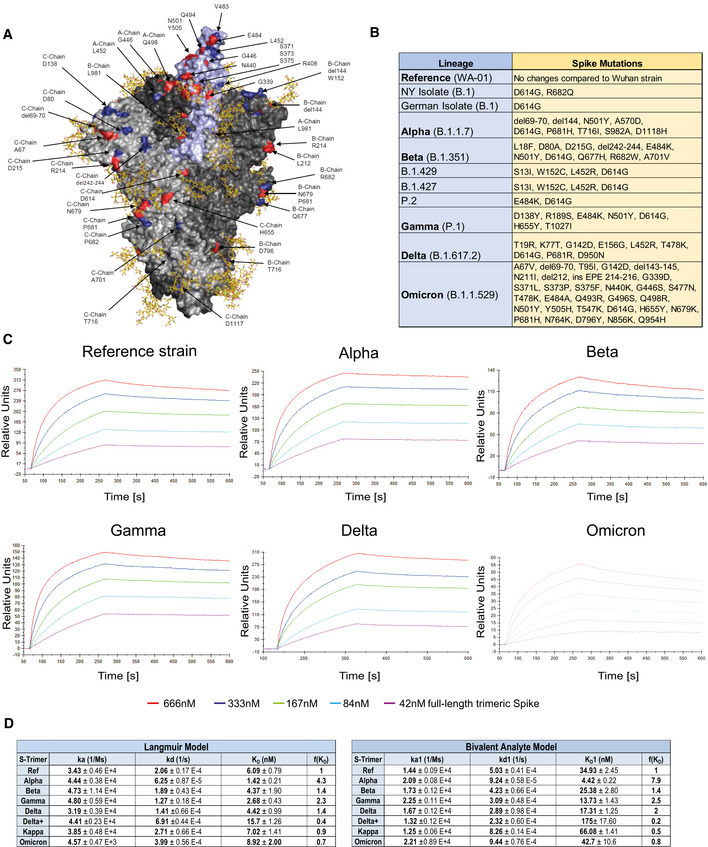
Figure 2. Increased binding affinity of APN01 to full‐length pre‐fusion trimeric Spike proteins from SARS‐CoV‐2 variants of concern.
-
APyMOL rendering of the trimeric full‐length SARS‐CoV‐2 Spike protein. One RBD is shown in violet. Indicated in blue are positions mutated in the various strains of SARS‐CoV‐2 used in experiments in this study. Mutations described for the Omicron VOC are depicted in red. Shown in yellow are the glycan modifications of the spike protein (Capraz et al, 2021).
-
BTable lists the SARS‐CoV‐2 strains and their respective mutations within the Spike protein that were used in this study.
-
CRepresentative sensorgram images for the SPR analysis conducted with full‐length trimeric spike proteins in pre‐fusion state with APN01. Reference strain corresponds to original Wuhan viral isolate spike sequence. Indicated are VOC Alpha, Beta, Gamma, and Delta, and Omicron.
-
DTables listing ka, kd, as well as KD values for the interaction of APN01 and full‐length trimeric spike proteins. The constants represent mean values and standard deviations obtained from sensorgram fittings performed in quadruplicate. Values are derived from calculations based upon the Langmuir (left table) or Bivalent Analyte sensorgram fitting (right table).
