Figure 3.
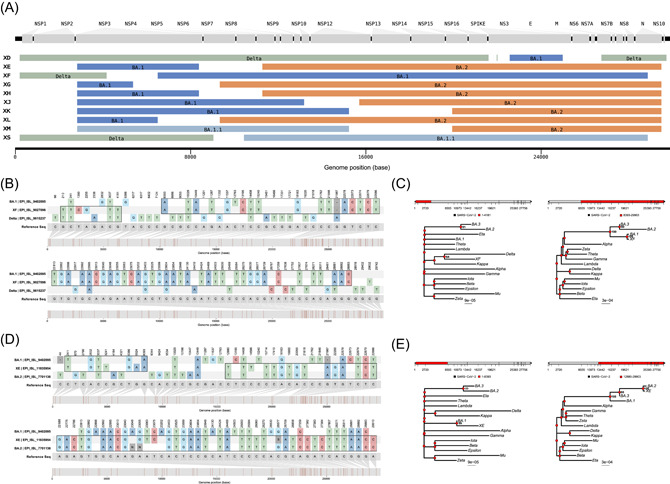
Pairwise nucleotide comparison and phylogenetic analysis of Delta–Omicron XF and XE. (A) Sequences of putative recombinants raised by Pango nomenclature exhibit the mosaic genomes structures composed of parental lineages. Tracts colored green, dark blue, pale blue, and orange represent the lineage Delta, BA.1, BA.1.1, and BA.2, respectively. Gaps represent the vague positions for recombinant breakpoints, there are no lineage‐specific mutations within these regions. The parental lineages for each putative recombinant are noted on the corresponding bar. (B) Pairwise nucleotide comparison of Delta, XF, and Omicron BA.1 against the reference sequence (Wuhan‐Hu‐1). (C) Phylogenetic analysis of different portions of the reported sequence of XF has been compared with VOCs, VOIs, and Omicron subvariants. (D) Pairwise nucleotide comparison of XE, Omicron BA.1, and BA.2 against the reference sequence (Wuhan‐Hu‐1). (E) Phylogenetic analysis of different portions of the first reported sequence of XE has been compared with VOCs, VOIs, and Omicron subvariants. VOCs, variants of concerns; VOIs, variants of interest.
