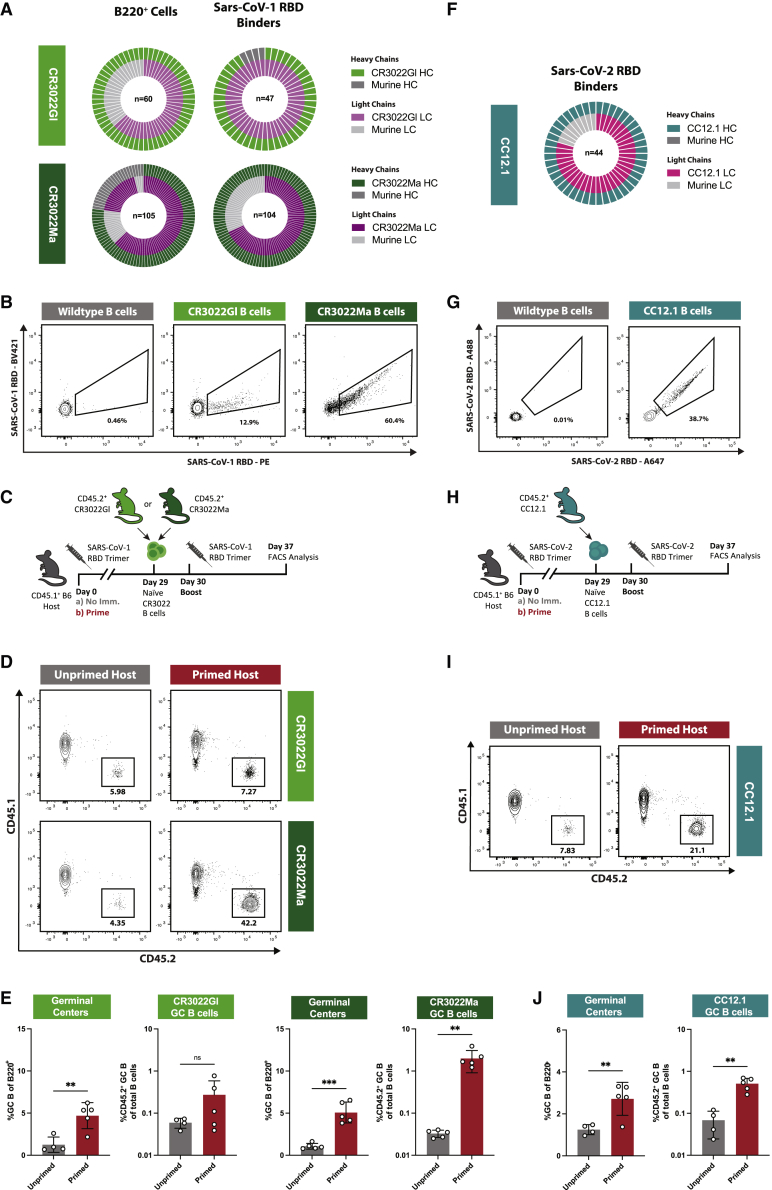
Figure 2.
Naive CR3022Ma but not CR3022Gl B cell responses are enhanced in primed mice
(A) (Top) Nested pie chart of CR3022Gl HC (light green), murine HC (dark gray), CR3022Gl LC (light purple), and murine LC (light gray) sequences amplified from single-cell sorted B220+ naive B cells from two CR3022GL mice. n = sequence pairs amplified. (Bottom) As top, for CR3022Ma.
(B) Representative FACS plot of binding of naive CR3022Gl and CR3022Ma and WT B cells to SARS-CoV-1 RBD probes.
(C) Schematic of evaluation of the effect of previous exposures on naive CR3022Gl and CR3022Ma B cell responses.
(D) Representative FACS plots of gating strategy to quantify GC B cell responses as the percentage of total B cells and the percentage of CD45.2+ (here CR3022Gl/Ma) GC B cells of the total GC B cell population.
(E) (Left) GC cells as the percentage of total B cells and (left-center) CR3022Gl GC B cells as percentage of total B cells at experiment day 40 (10 dpi) in previously unprimed (gray) or primed (red) CR3022Gl recipients. (Right-center) GC cells as the percentage of total B cells and (right) CR3022Ma GC B cells as the percentage of total B cells at experiment day 40 (10 dpi) in previously unprimed (gray) or primed (red) CR3022Ma recipients.
(F) Nested pie chart of CC12.1 HC (teal), murine HC (dark gray), CC12.1 LC (pink), and murine LC (light gray) sequences amplified from single-cell sorted B220+ naive B cells from 2 CC12.1 mice. Center number indicates sequence pairs amplified.
(G) Representative FACS plot of binding of naive CC12.1 and WT B cells to SARS-CoV-2 RBD probes.
(H) Schematic of experimental evaluation of the effects of previous exposures on naive CC12.1 B cell responses in primed and unprimed recipients.
(I) Representative FACS plots of gating strategy to quantify GC B cell responses as the percentage of total B cells and the percentage of CD45.2+ (here CC12.1) GC B cells of the total GC B cell population.
(J) (Left) GC cells as the percentage of total B cells and (right) CC12.1 GC B cells as the percentage of total B cells at experiment day 37 (7 dpi) in previously unprimed (gray) or primed (red) CC12.1 recipients.
p values were calculated by unpaired Student’s t test (∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant). Figures represent data from one experiment with 4–5 mice per condition, with data presented as mean ± SD.
See also Figure S4.