Figure 1:
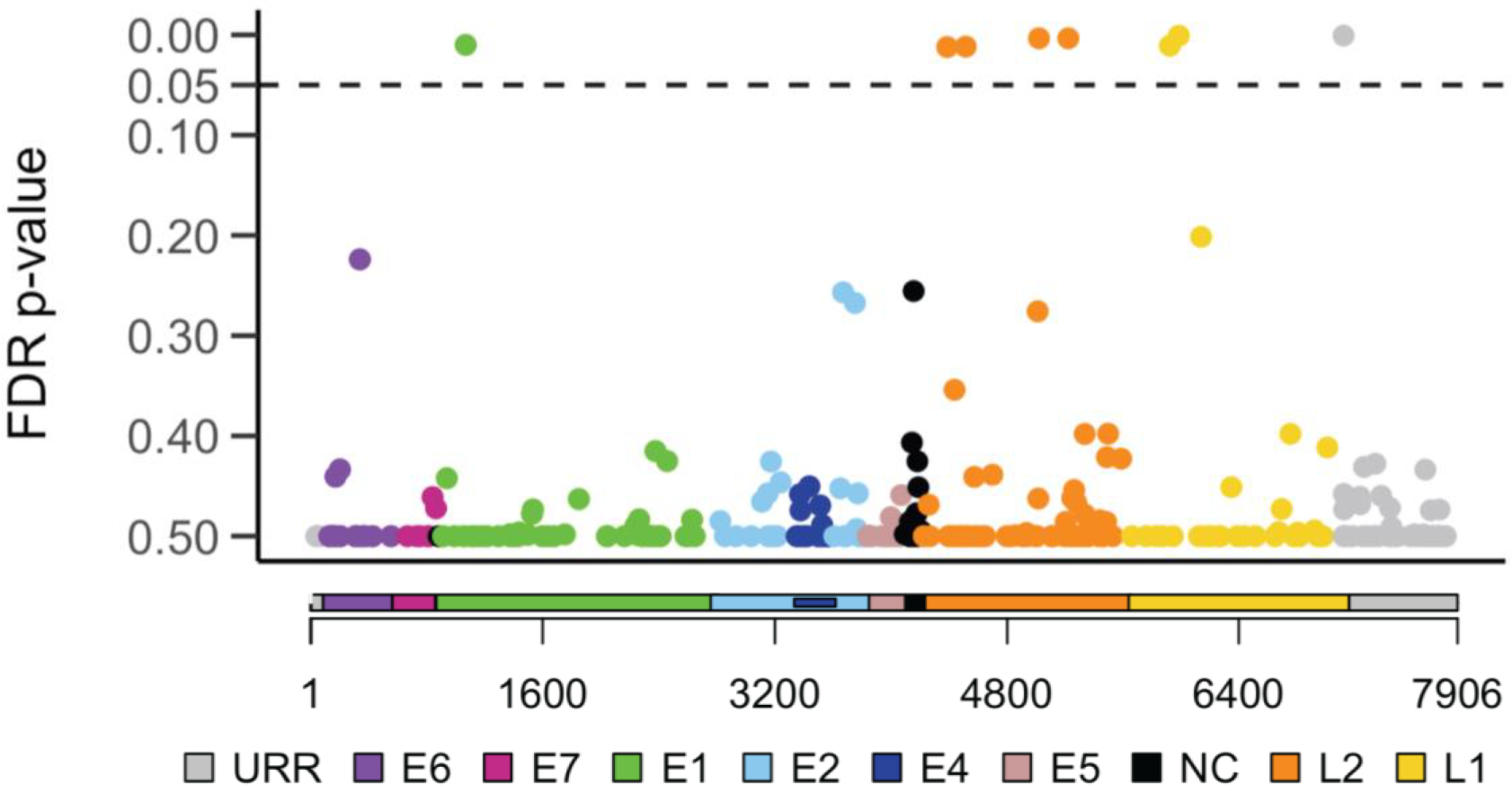
Individual HPV16 SNP associations with overall survival
Cox proportional hazards models were used to generate hazard ratios and 95% confidence intervals for the risk of death (overall survival) for each minor HPV16 SNP. The major allele at each SNP position was used as the referent group. A total of 284 SNPs varied between the 384 HPV16 genomes sequenced and were included in the analysis. The Y-axis denotes the false discovery rate (FDR) corrected Ps for the hazard ratio of each outcome. The X-axis denotes the position of each SNP in the HPV16 genome; each gene is identified with a different color. URR, upstream regulatory region (grey); E6, early gene 6 (purple); E7, early gene 7 (magenta); E1, early gene 1 (green); E2, early gene 2 (light blue); E4, early gene 4 (dark blue); E5, early gene 5 (pink); NC, non-coding region (black); L2, late gene 2 (orange); L1, late gene 1 (yellow). Dotted line denotes the FDR cutoff of 0.05.
