Figure 3. Overview of the design of antibody sub-libraries for affinity maturation.
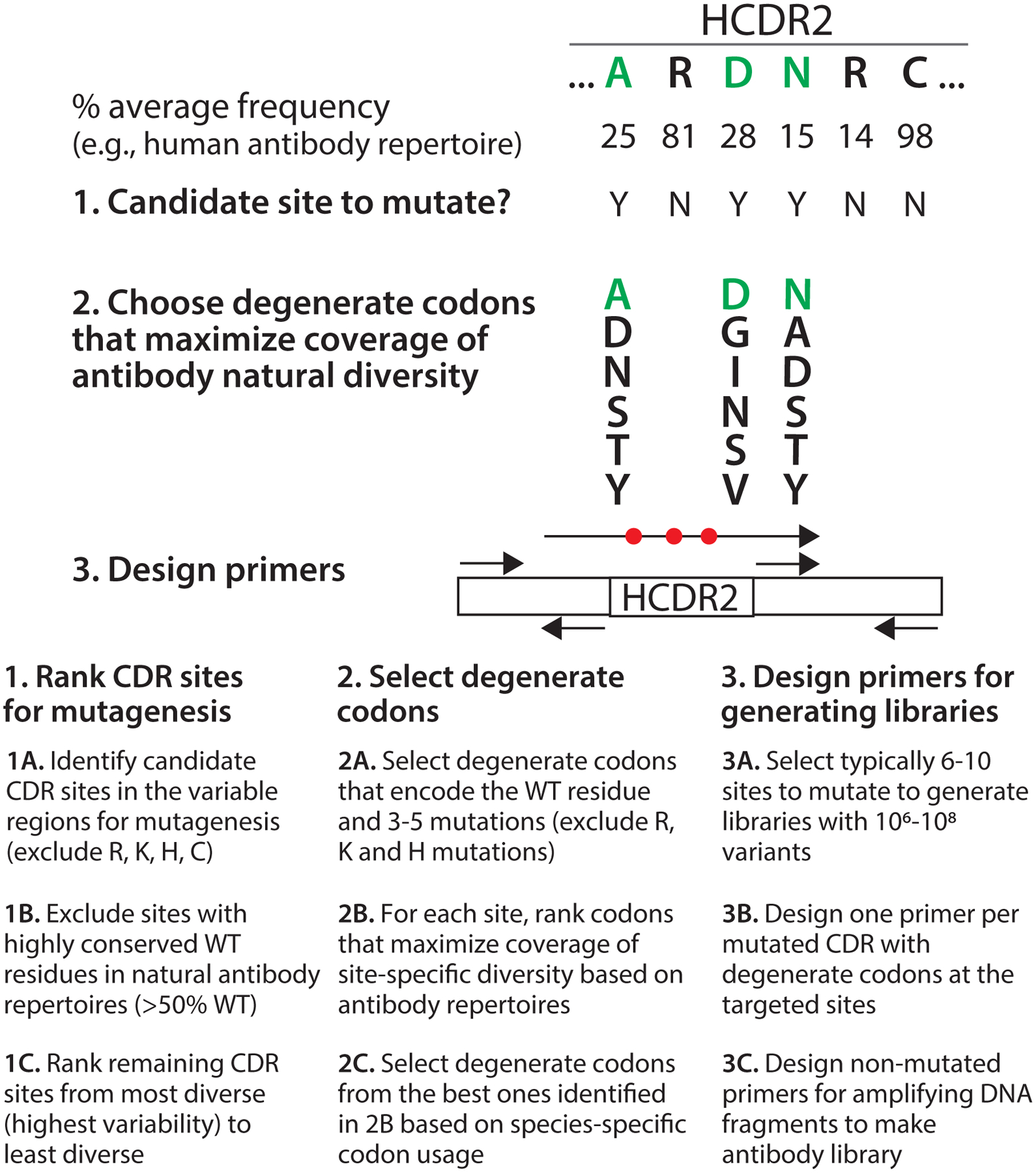
The design of sub-libraries for affinity maturation involves three major steps. First, the CDR positions to mutate are chosen based on the variability of each CDR position in natural antibody repertoires. The sites are ranked from most variable (most attractive for mutagenesis) to least variable (least attractive for mutagenesis), and highly conserved positions (>50% WT on average in natural antibody repertoires) are eliminated from consideration. Moreover, a subset of CDR sites are also excluded from consideration if their WT residues are Arg, Lys, His or Cys. Second, for the selected ~6–10 CDR sites with the highest variability, degenerate codons are chosen that encode the WT residue as well as 3–5 other amino acids based on maximizing the sum of the average site-specific frequencies of each encoded residue in natural antibody repertoires (referred to as natural diversity coverage). Degenerate codons with Arg, Lys and His are excluded. The libraries are designed to typically encode 106-108 variants. If there are multiple possible degenerate codons that encode the same set of amino acid mutations, codon selection is based on species-specific codon usage (e.g., S. cerevisiae codon usage). Third, mutagenic primers with degenerate codons and amplification primers without mutations are designed. One mutagenic primer is designed for each mutated CDR that contains the site-specific degenerate codons and 18–22 base pairs of framework DNA on both ends of the mutated CDR. Three additional primers are required for generating sub-libraries with a single mutated CDR. Typically two or three CDRs are mutated when generating sub-libraries for affinity maturation.
