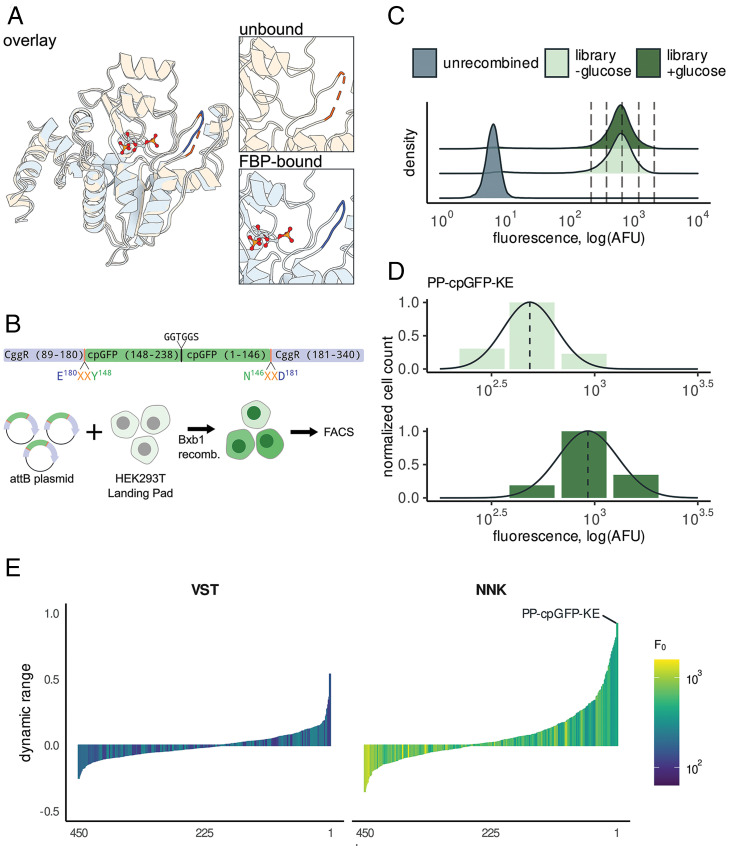
Fig. 1.
Discovery of a high dynamic-range FBP biosensor by sort-seq assay. (A) Structural comparison of CggR in apo state (PDB ID code 2OKG, orange) vs. FBP bound (PDB ID code 3BXF, blue) reveals a loop (residues 177 to 183) that undergoes a disorder-to-order transition upon binding FBP (6). (B) A library of linker variants were assayed for function in HEK293T Landing Pad cells. Circularly permuted GFP was inserted into CggR at residue 180 with two flanking linker amino acids on either side. Linker amino acids were encoded by the degenerate codon VST, which translates to a limited set of 8 amino acids, or by the fully degenerate codon NNK, which includes all 20 amino acids. The library was placed into an attB plasmid that enables genomic recombination into the HEK293T Landing Pad genome by the Bxb1 recombinase. (C) Fluorescence distributions of the CggR-180-NNK library expressed in HEK293T cells exposed to 0 mM or 25 mM glucose. Dotted lines indicate the bins used for sort-seq. (D) The number of cells sorted into each bin is indicated by bar height along with the maximum-likelihood density estimates for the variant with the highest dynamic range, CggR-180-PP-cpGFP-KE (HYlight). (E) Dynamic-range (ΔF/F) estimates for all variants after screening libraries with linker residues substituted with amino acids encoded by a limited set using VST codons (Left) or fully degenerate NNK codons (Right). See also SI Appendix, Fig. S1.