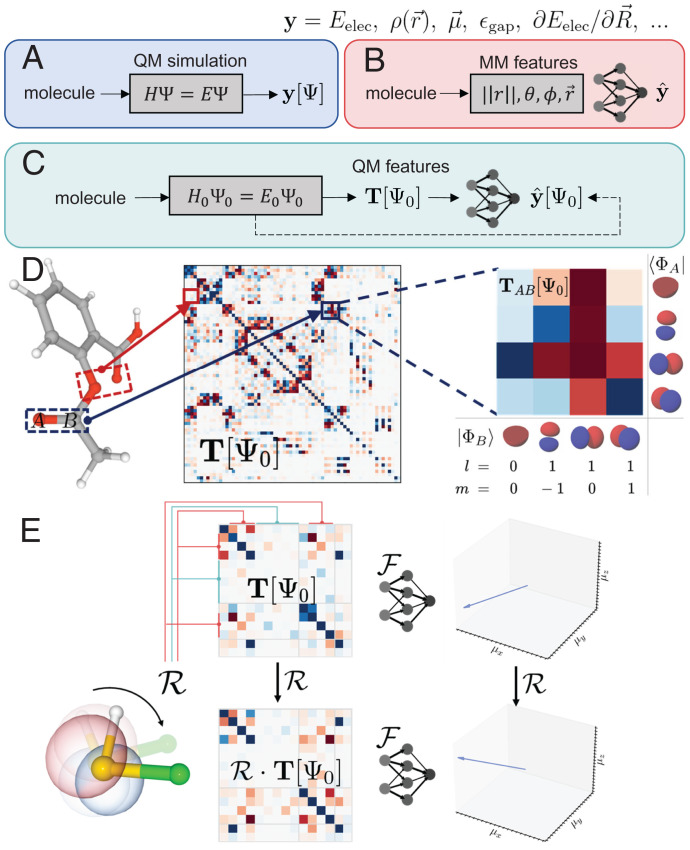
Fig. 1.
QM-informed ML for modeling molecular properties. (A) Conventional ab initio quantum chemistry methods predict molecular properties based on electronic structure theory through computing molecular wave functions and interaction terms, with general applicability but at high computational cost. (B) Atomistic ML approaches use geometric descriptors, such as interatomic distances, angles, and directions, to bypass the procedure of solving the electronic structure problem but often require vast amounts of data to generalize toward new chemical species. (C) In our approach, features are extracted from a highly coarse-grained QM simulation to capture essential physical interactions. An equivariant neural network efficiently learns the mapping, yielding improved transferability at an evaluation speed that is competitive to Atomistic ML methods. (D) Characteristics of the atomic orbital features considered in OrbNet-Equi. The features T are visualized by the density matrix of the molecular system, with red color indicating positive matrix elements and blue color indicating negative matrix elements. Every pair of atoms (A, B) is mapped to a block in the feature matrix, with the row dimension of the block matching the atomic orbitals of the source atom A and the column dimension matching the atomic orbitals of the destination atom B. (E) OrbNet-Equi is equivariant with respect to isometric basis transformations on the atomic orbitals (Eqs. 3 and 4), yielding consistent predictions (illustrated as the dipole moment vector of a thiohypofluorous acid molecule) at different viewpoints. Pink and blue densities overlaying the molecule illustrate an atomic orbital; upon viewpoint rotation , the original atomic orbital (semitransparent) is reexpressed as a linear combination of atomic orbitals in the rotated frame, leading to a basis transformation that changes the coefficients of feature matrices T.