Fig. 4.
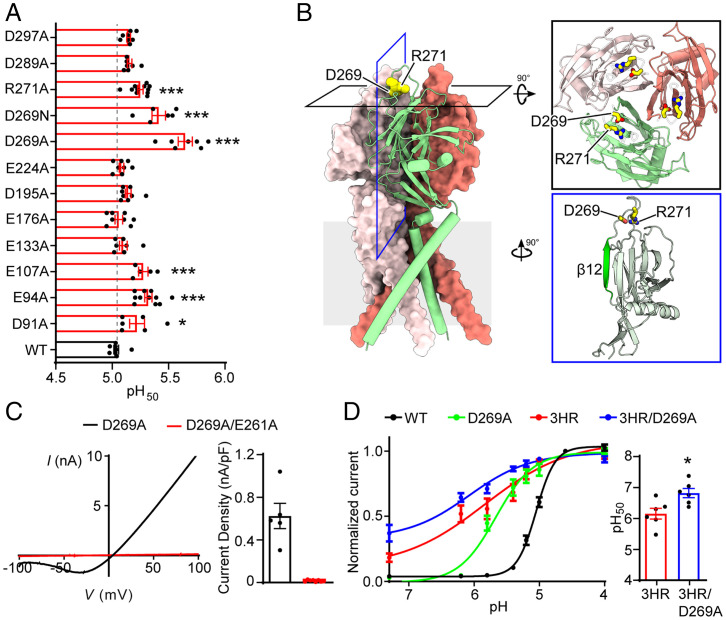
Screening of acidic residues in the ECD identifies D269 as another regulator of PAC pH sensitivity. (A) pH50 values (mean ± SEM) of wild-type PAC and mutants. *P < 0.05; ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (B) Cryo-EM structure models of pH 4–PAC structure viewed parallel to the membrane (Protein Data Bank ID: 7JNC). The green subunit is shown as a cartoon and the other two subunits are shown in surface representation. D269 and R271 are highlighted as yellow spheres. Inserts: Top view (black frame) and side view (blue frame) highlighting D269 and R271. The β12 strand is highlighted in green in the side view. (C) Left: Representative whole-cell currents of D269A (black) and D269A/E261A (red) mutants at pH 4.6, and monitored by voltage-ramp protocol. Right: pH 4.6–induced current densities (mean ± SEM) of D269A and D269A/E261A mutants at +100 mV. (D) Left: The pH dose-response curve of wild-type PAC (black), D269A (green), 3HR (red), and 3HR/D269A (blue). The currents are normalized to the pH 4.0–induced currents [n = 7 (wild-type PAC); n = 7 (D269A); n = 6 (3HR); n = 6 (3HR/D269A)]. The normalized data are fitted to the Hill equation unconstrained. Data are the mean ± SEM of the normalized currents at +100 mV. Right: pH50 values (mean ± SEM) of 3HR and 3HR/D269A mutants estimated from the pH dose-response curve in (D). *P < 0.05, two-tailed Student’s t test.