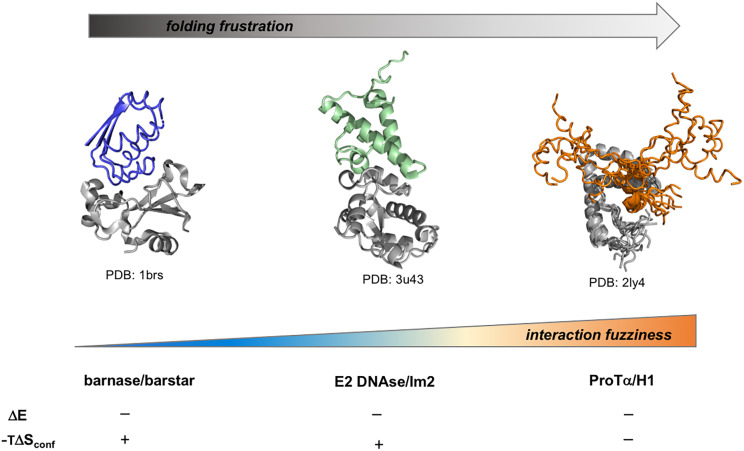
Fig. 1.
Entropy modulates the affinity of ordered and disordered complexes. High-affinity binding can be achieved through both structured and disordered interfaces. An ordered complex is represented by the ribonuclease barnase and its inhibitor barstar [blue, Protein Data Bank (PDB) code 1brs (25)]. A partially ordered complex is represented by the endonuclease colicin E2 with its cognate immunity protein [Im2, green, PDB code 3u43 (26)]. A disordered complex is represented by the complex between HMGB1 and p53 [orange, PDB code 2ly4 (27)], as the high-resolution structure of the ProTα/H1 complex is not available. Increasing folding frustration decreases structural order from barnase/barstar to HMGB1/p53 complex (from left to right, top arrow). Following a similar trend, frustration of the interface contacts also increases from ordered (blue) to disordered (orange) complexes. A high degree of frustration results in a conformationally heterogeneous complex and positive change in configurational entropy (TΔSconf). Electrostatic interactions provide major contributions to binding affinity in all these scenarios. While charged residues in the ordered complex are engaged in specific contacts, in disordered complexes they modulate the degree of disorder and binding entropy (4, 27).