Fig. 5.
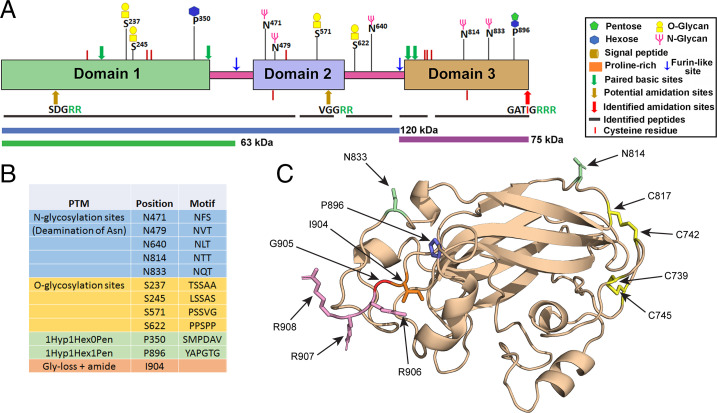
Posttranslational modification of HEK-proGATI. (A) Peptides identified by mass spectrometry (black bars) cover most of the three stable domains (domains 1, 2, and 3 are shown) and the linker regions of proGATI; the regions predicted to correspond to the 120-, 75-, and 63-kDa products are indicated by colored bars. The symbols used to indicate the various modifications are shown. All C-terminal peptides had been proteolytically processed and amidated; C-terminal peptides ending in –Gly, –Gly-Arg, –Gly-Arg-Arg, or –Gly-Arg-Arg-Arg were not found. (B) Table indicating the posttranslational modifications identified. (C) Detailed AlphaFOLD 2 structural prediction for domain 3. The two disulfide bonds, glycosylated Asn and HyP residues are shown. The C-terminus has three Arg residues (R906-R908) that are proteolytically removed. The exposed Gly residue (Gly905) is then α-hydroxylated by the PHM domain of PAM and cleavage by its lyase domain releases glyoxylate and generates a C-terminal Ile904-amide.