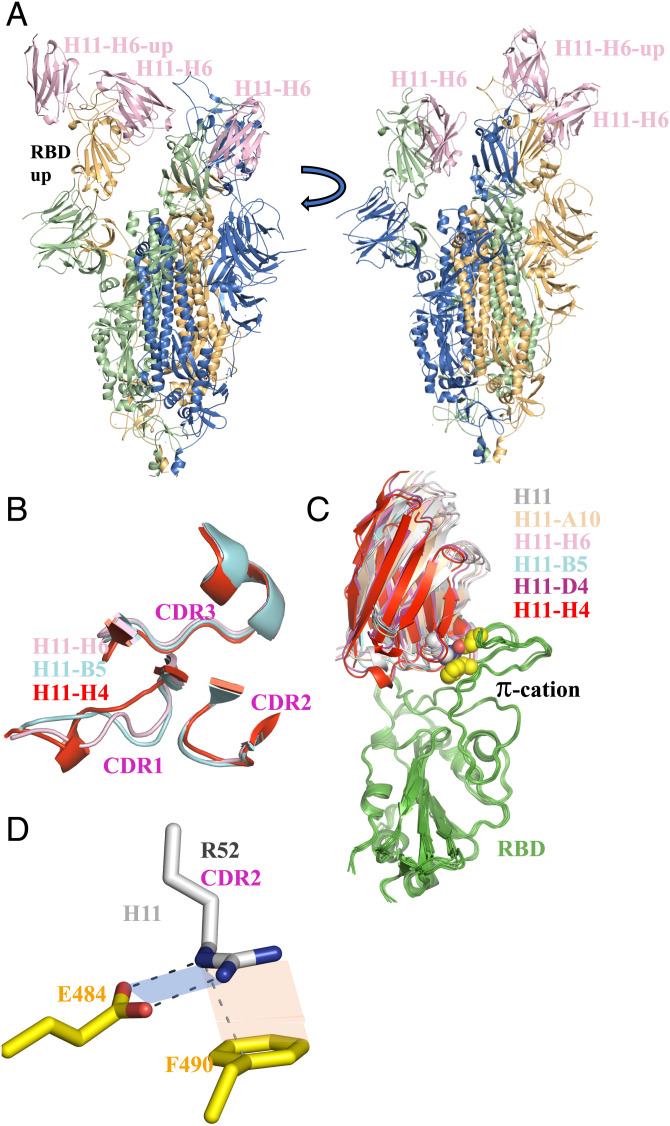
Fig. 1.
Cryo-EM structures of the nanobodies bound to the spike protein of SARS-CoV-2. (A) Cryo-EM structure of a nanobody (H11-H6) bound to spike. Nanobodies H11, H11-A10, and H11-B5 all give the same one-up–two-down conformation of the spike which was first described for nanobodies H11-H4 and H11-D4 (29). A second view (rotated 90° around the vertical axis) is shown. The sequence variation in the nanobodies is confined to a six-residue segment of the CDR3 loop (Table 1). (B) Although the residues that vary between the six nanobodies are located in the CDR3 region, structural changes are actually seen in CDR1. Shown are the three nanobodies which illustrate the different CDR1 structures observed. (C) Superposition of the RBD-nanobody complexes reveals the nanobodies bind in slightly different orientations on the surface of the RBD. The RBD molecule is colored green, and nanobodies are colored individually as follows: H11 (gray), H11-A10 (wheat), H11-H6 (light pink), H11-B5 (cyan), H11-D4 (purple), and H11-H4 (red). The complexes are anchored by the π–cation involving R52 from CDR2 of the nanobody (the side chain as spheres with carbon atoms colored white, nitrogen blue) interacting with E484 and F490 (carbon atoms yellow, oxygen atoms red) from RBD. (D) Close-up view of the π-cation interaction from the H11 complex. The geometry (distance and orientation) of both the salt bridge (blue plane) and the π stacking interaction (orange plane) varies the complexes (Table 2).