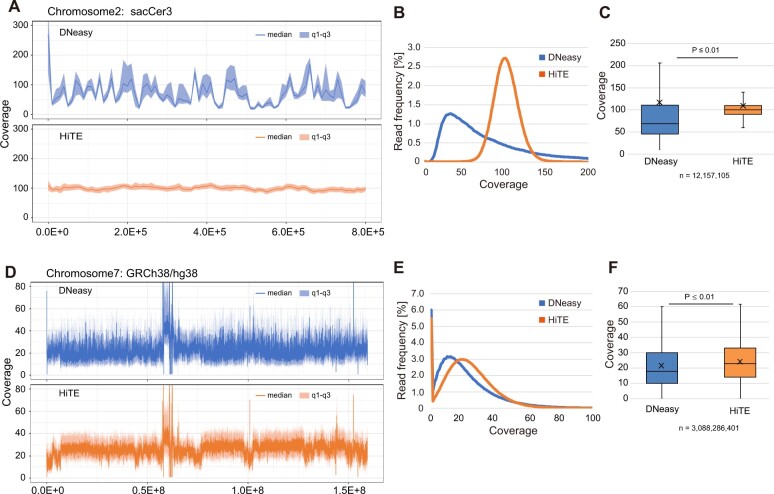
Figure 4:
HiTE produces reads that are more evenly mappable to the reference genome. (A–C) DNA recovered from fixed budding yeast nuclei with DNeasy and HiTE are compared. (D–F) Comparison of DNA extraction from scratched tumor cells in a retinoblastoma tissue section (Supplementary Fig. S3) using DNeasy and HiTE. The representative genome browser shot (A and D), the distribution (B and E), and box plot (C and F) of mapped read coverage are shown. P-values were calculated using Welch’s t-test (C and F).