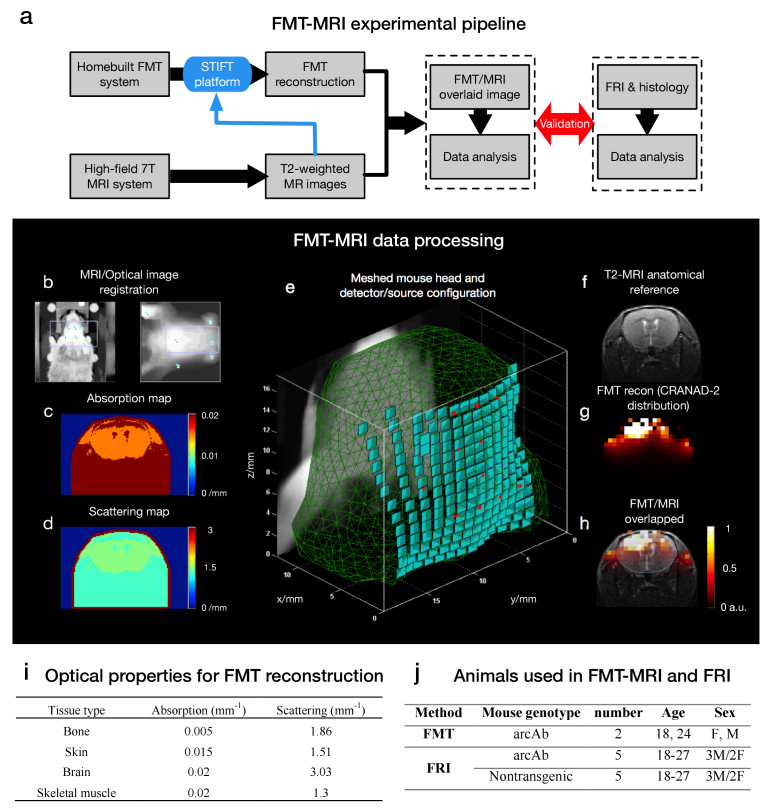
Fig. 1.
The multimodal FMT-MRI experimental pipeline and data processing framework are illustrated in the upper (a) and middle (b-h) panels, respectively. Regarding the framework of FMT-MRI data processing, the topological surface of anatomical MRI (b, right) was first registered with the optical image (b, left) with fiducial markers indicated on both images (blue dot). Once the two types of images were registered, both an absorption map (c) and a scattering map (d) were generated according to the intensity-based segmentation. (e) MRI-derived structural information was used to reconstruct the object in 3D, and adaptive meshing was performed with detector elements (green patches) and laser illumination points (red spots) assigned to the surface of the generated mesh. (f) T2-weighted MR image (f) provides an important anatomical reference that can significantly improve the localization of the FMT signal (g). (h) shows an overlaid FMT-MRI image containing both molecular and structural information within the same mouse. (i) The absorption and scattering coefficients of four typical tissue categories (bone, skin, brain and skeletal muscle) were used for FMT reconstruction. (j) Summary of animals used in FMT-MRI and FRI experiments.