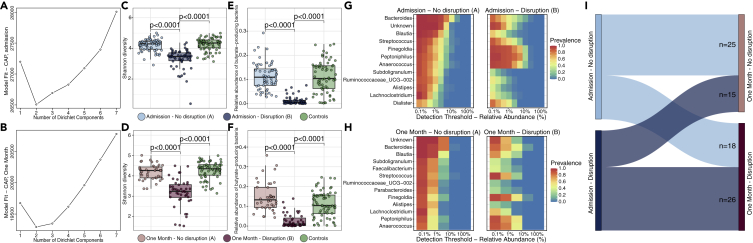
Figure 2.
Unsupervised clustering of microbiota profiles during and following CAP
Laplace approximation of model fitting showed that two clusters offered the best fit of patients at admission (A) and after one month (B): one with disrupted microbiota (n = 55 at admission; n = 44 after one month) and the other with undisrupted microbiota (n = 60 at admission; n = 40 after one month). Cluster B (“disruption”) had significantly decreased alpha diversity compared to cluster A (“no disruption”) and controls, both at admission (C) and after one month (D), as well as lower relative abundances of butyrate-producing bacteria in the disrupted cluster at admission (E) and after one month (F). The undisrupted cluster had high prevalence and relative abundance of Bacteroides and members of the Ruminococcaceae and Lachnospiraceae families (G), while the disrupted cluster was dominated by facultative anaerobic peptococci (e.g. Finegoldia, Peptoniphilus, and Anaerococcus) (H). Sankey diagram showing the number of patients (represented by the size of the links) that remained within the same cluster between the two timepoints, or shifted between microbiota clusters (e.g. from the undisrupted cluster at admission to the disrupted cluster one month thereafter) (I). In the boxplots, the central rectangle spans the first quartile to the third quartile (the interquartile range or IQR), the central line inside the rectangle shows the median, and whiskers above and below the box. Given the non-parametric nature of the data, p values were calculated using the Wilcoxon rank-sum test. In the heatmap, the color of each cell shows the percentage of samples (prevalence) in which a specific bacterial genus (y axis) is present at different relative abundances (x axis).