Figure 3.
Relating gut microbiota to cytokine responses during and following CAP hospitalization
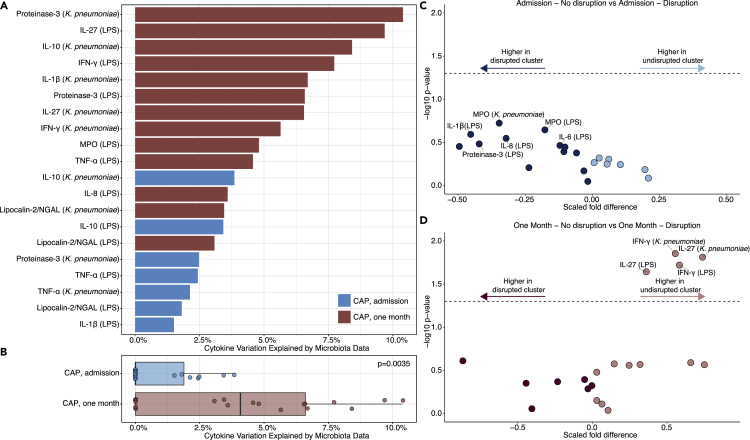
(A) Rectal microbiota accounted for a maximum of 10.4% of interindividual variation in cytokine production capacity of CD14+ monocytes and polymorphonuclear cells upon ex vivo stimulation with LPS or heat-killed K. pneumoniae.
(B) Considerably less variation in cytokine responses was explained by the microbiota at hospital admission for CAP compared to at one month following hospitalization. Microbiota are represented by the first four principal coordinates (PCoA with weighted Unifrac distance) that explain ∼65% of the variance, to avoid overestimation due to species-species correlations. The cytokine variance explained by these principal coordinates was estimated through permutation ANOVA by summing over the significant contributions (p < 0.2). In the boxplot, dots represent the percentage of cytokine variation explained by microbiota data for each cytokine response (also shown in panel A), the central rectangle spans the first quartile to the third quartile (the interquartile range or IQR), the central line inside the rectangle shows the median, and whiskers above and below the box.
(C) No significant differences (p > 0.05) in cytokine responses between clusters A (n = 55; undisrupted) and B (n = 60; disrupted) at hospital admission.
(D) One month following hospitalization, monocytes of patients with undisrupted microbiota profiles (n = 40) produced higher levels of IFN-ɣ and IL-27 compared to patients with disrupted microbiota profiles (n = 44). Given the non-parametric nature of the data, p values were calculated using the Wilcoxon rank-sum test.