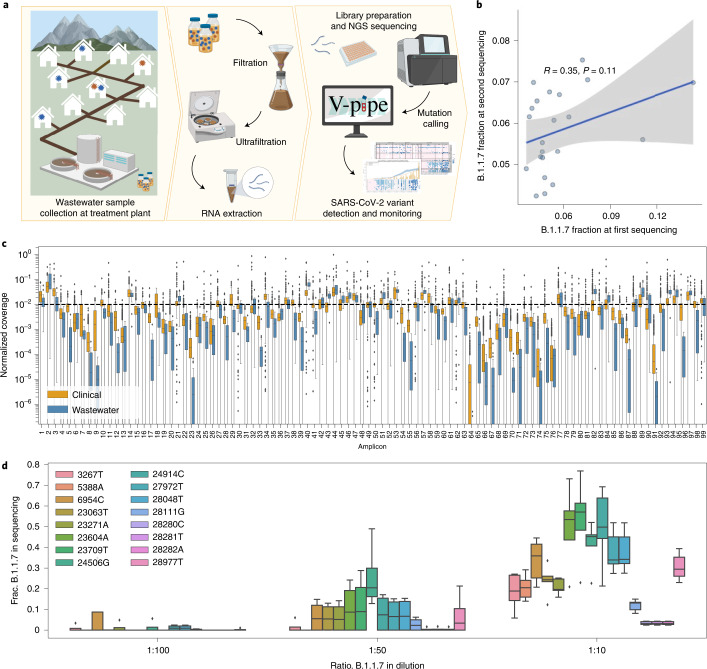
Fig. 1. Method overview and quality control.
a, Overview of the wastewater sampling campaign. Left: collection of raw wastewater samples containing a mixture of wild-type and variant SARS-CoV-2 viral RNA. Middle: viral concentration and nucleic acid extraction. Right: amplification using ARTIC v3 primers, library preparation, NGS and mutation calling using V-pipe, followed by statistical analysis to detect and quantify the presence of SARS-CoV-2 variants and estimate epidemiological parameters. Created with BioRender.com. b, Reproducibility of Alpha (B.1.1.7) prevalence based on resequencing of 25 samples. Each dot shows the average fraction of Alpha-compatible reads across all signature mutations. Pearson correlation coefficient, R, and P value (two-sided test) indicate a high degree of variability in Alpha prevalence estimates at low frequencies. The solid line denotes the estimate from the linear model and the shaded area denotes the 95% confidence interval. c, Per-amplicon normalized coverage distributions after quality filtering and alignment in the same NGS batch containing both 589 clinical (orange) and 22 wastewater (blue) samples. Per-amplicon absolute coverages can be found in Supplementary Fig. 1. d, Reproducibility of Alpha (B.1.1.7) prevalence in a dilution series experiment. Boxplots represent fractions of substitutions called in 5 technical replicates of wastewater spiked with SARS-CoV-2 RNA at 3 different Alpha-to-wild-type ratios. In both c and d, boxes show quartiles and the whiskers extend to a maximum of 1.5× the interquartile range, after which points are considered outliers.