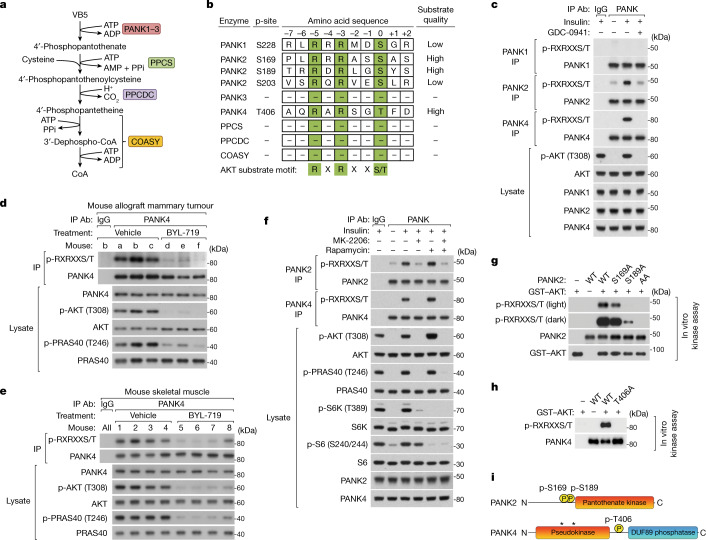
Fig. 2. PANK2 and PANK4 are direct AKT substrates.
a, CoA synthesis pathway enzymes. The full names and accession numbers are provided in Supplementary Table 1. b, Enzymes of the CoA synthesis pathway that are candidate AKT substrates. Amino acid residues that fall within low- to high-quality AKT substrate motifs according to the kinase–substrate prediction program Scansite, and that are reported to be phosphorylated in the phosphoproteomic database Phosphosite are listed. Numbering is based on the human sequence. c, Endogenous PANK1, PANK2 and PANK4 immunoprecipitation (IP) from MCF10A cells with insulin (100 nM) and PI3K inhibitor (GDC-0941, 2 μM) treatments (30 min), preceded by serum and growth factor deprivation (18 h) and inhibitor pretreatment (15 min). d, Endogenous PANK4 immunoprecipitation from orthotopic mammary allograft tumours in C57BL/6J treated with vehicle or PI3K inhibitor (BYL-719, 45 mg kg−1) daily for 10 days. e, Endogenous PANK4 immunoprecipitation from skeletal muscle (gastrocnemius) of C57BL/6J mice treated with PI3K inhibitor (BYL-719, 50 mg kg−1) for 1 h. f, Endogenous PANK2 and PANK4 immunoprecipitation from MCF10A cells treated with insulin (100 nM), AKT inhibitor (MK-2206, 2 μM), and mTORC1 inhibitor (rapamycin, 20 nM) with conditions otherwise as in c. g,h, In vitro AKT kinase assays. Untagged PANK2 (g) or PANK4 (h) (WT or alanine point mutants) were immunopurified from respective reconstituted knockout cells treated with PI3K and AKT inhibitors. PANK immunopurifications were incubated with purified GST–AKT (30 min). i, Diagram of the human PANK2 and PANK4 domains and AKT-targeted phosphorylation sites. The asterisks indicate the location of evolutionary mutations inactivating PANK4 kinase domain. For c–h, immunoblotting analysis probed for total and phosphorylated proteins including AKT phospho-substrate motifs; representative of two independent experiments. IgG, control IgG immunoprecipitation.