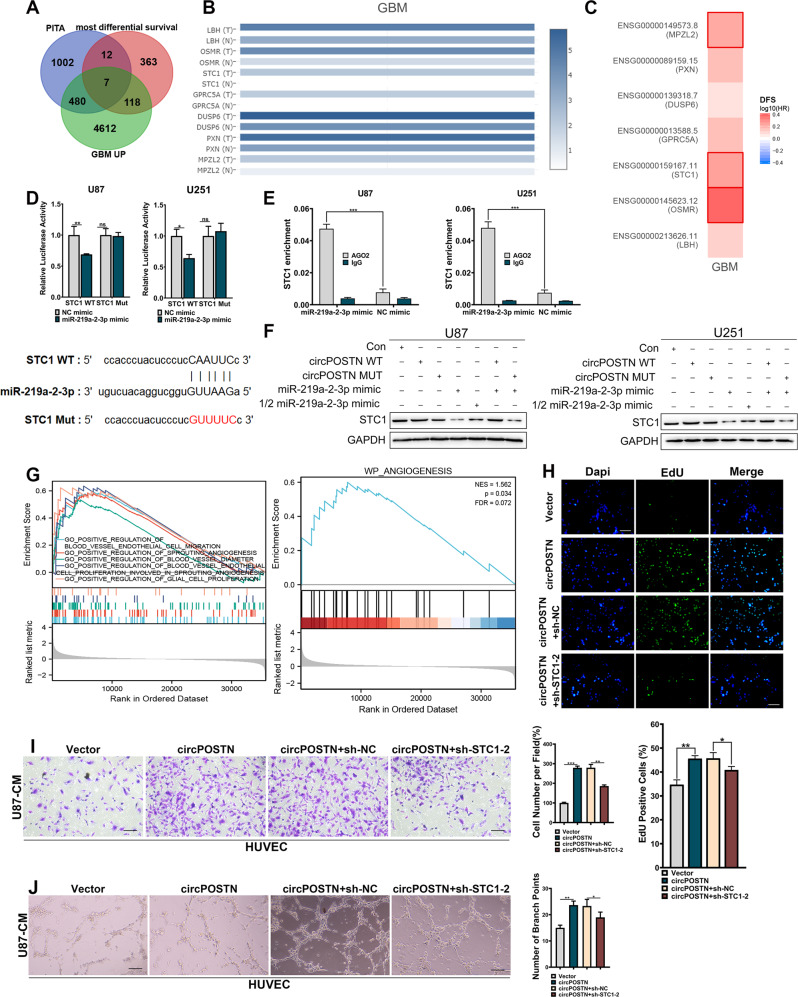
Fig. 5. CircPOSTN acts as a ceRNA and binds with miR-219a-2-3p to upregulate STC1 expression.
A The intersection of genes predicted to: i) binds with miR-219a-2-3p based on an online tool PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html), n = 1501; ii) be the most differential survival genes in GBM tissues based on GEPIA2 database, n = 500; iii) be upregulated in GBM tissues based on GEPIA2 database, n = 5217. Seven genes (LBH, OSMR, STC1, GPRC5A, DUSP6, PXN, and MPZL2) were identified. B The expression levels of these seven genes in GBM were analyzed based on the TCGA dataset. C The disease free survival (DFS) analysis of the relationship between these seven genes and GBM survival prognosis based on GEPIA2 database. D The sequence in the STC1 gene predicted to bind with miR-219a-2-3p was cloned into the luciferase reporter vectors (STC1 WT). The region-mutant sequence was used as control (STC1 Mut). Then, the dual-luciferase reporter assays were conducted to examine the relative luciferase activity of U87 and U251 cells co-transfected with STC1 WT-Mut vectors and miR-219a-2-3p mimic/control. E The expression level of STC1 was precipitated by anti-AGO2 antibodies using RIP assays. The U87 and U251 with miR-219a-2-3p overexpressed were used. F Western blotting analysis of STC1 in U87 and U251 cells co-transfected with STC1 WT-Mut vectors and miR-219a-2-3p mimic/control. The GAPDH was used as endogenous control. G GSEA results based on the TCGA dataset indicated that the high expression of STC1 was significantly correlated with the neovascularization and cell proliferation abilities of GBM. H–J The HUVECs were cultured in the conditioned medium from U87 cells with circPOSTN overexpressed with/without STC1 silenced. H Edu staining assay was used for measuring the proliferation ability of HUVECs. I The migration ability of HUVECs was detected by transwell migration assays. J The effects of circPOSTN and STC1 on the neovascularization ability of U87 cells were determined by tubule formation assays. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. NS not significant.