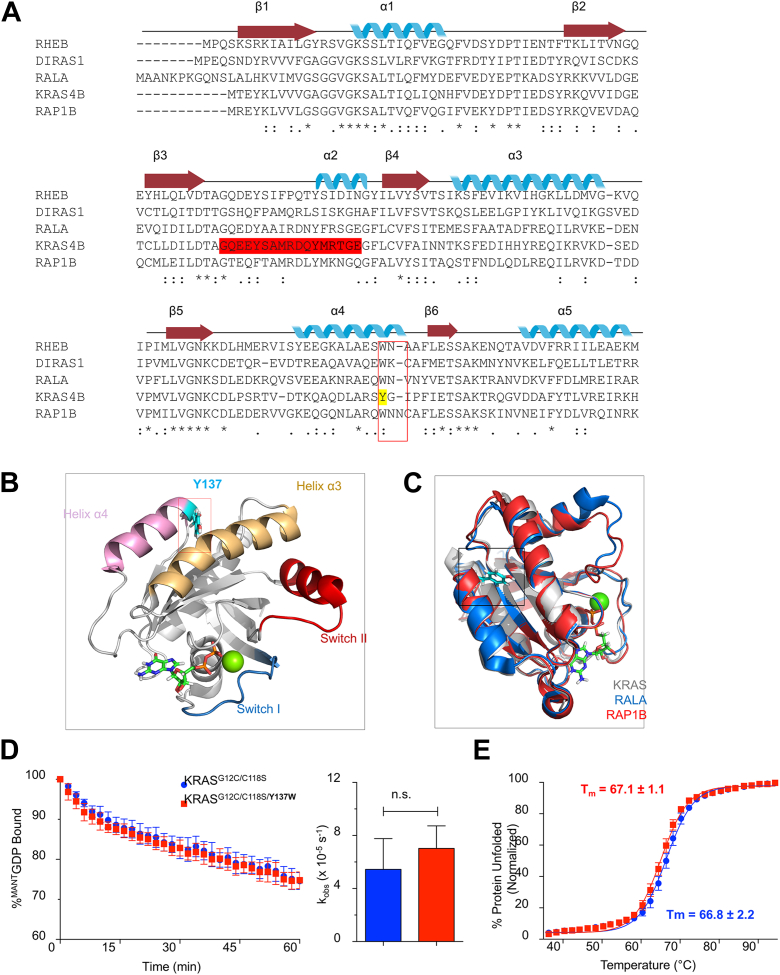
Figure 2.
Characterization of the Y137W substitution in the C118S variant of KRASG12C.A, sequence alignment of Ras subfamily GTPases showing conservation in the region of KRAS Y137. Alignment shows key alpha-helix and beta-sheet structural motifs (KRAS residues 1–165). The red box highlights the location of Y137 in KRAS and the corresponding tryptophan residue present in related family members. B, 3D location of residue Y137 in KRAS (PDB: 4LPK). Y137 is shown in sticks and highlighted in cyan, bound GDP is shown in sticks, and a bound calcium ion is shown in green. Switch I: teal, Switch II: red. Ribbon diagram was rendered using PyMOL. C, structural overlay of GDP-bound KRAS4B (PDB: 4LPK), RAL A (PDB: 6P0J), and RAP1B (PDB 3X1W) highlighting location of the residues equivalent to KRAS Y137 (black box). D, representative intrinsic nucleotide dissociation monitoring the decrease in mantGDP fluorescence over time after mixing with nonfluorescent GDP, comparing KRASG12C/C118S with and without the Y137W mutation, shows minimal effects of the Y137W mutation on nucleotide exchange activity. Data shown are representative from three or more independent experiments. Error bars, mean ± s.e.m. E, comparison of protein stability by CD thermal denaturation for KRASG12C/C118Sversus KRASG12C/C118S/Y137W. The similar midpoint Tm values indicate that Y137W does not notably impact protein thermal stability. Data shown are representative from three or more independent experiments. Error bars, mean ± s.e.m. CD, circular dichroism.