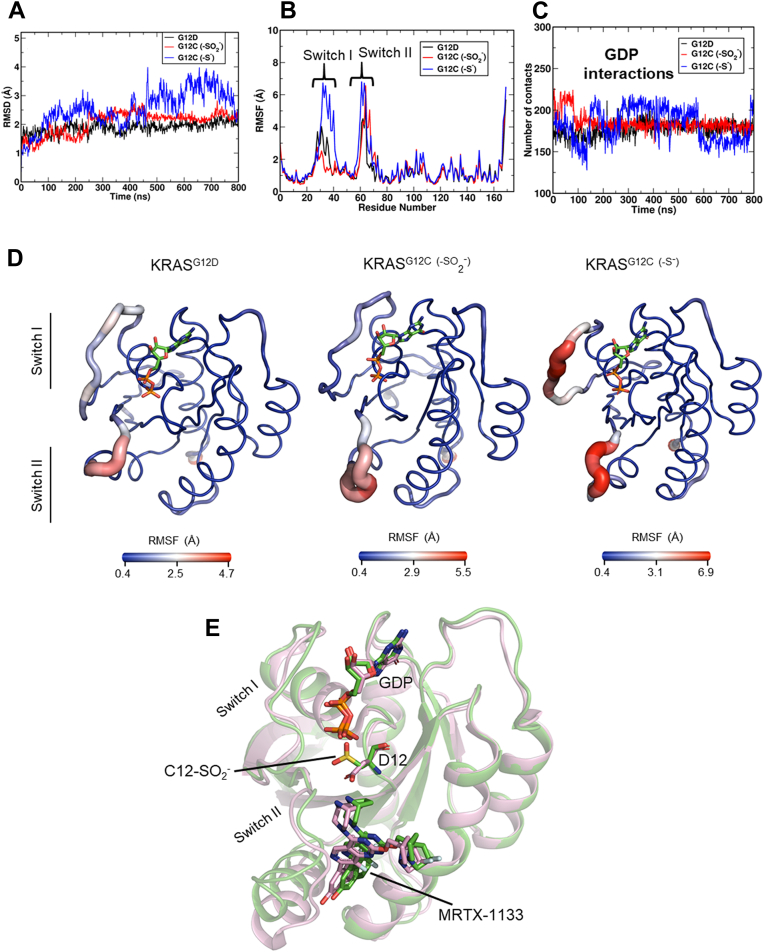
Figure 7.
KRASG12C(-SO2-) and KRASG12Dexhibit similar structural ensembles. Shown are (A) RMSD and (B) RMSF plots for GDP-bound KRASG12D, KRASG12C (-SO2-), and KRASG12C (-S-). C, quantification of atomic interactions for RAS-bound GDP with surrounding residues in KRASG12D, KRASG12C (-SO2-), and KRASG12C (-S-) indicate very similar GDP-binding interactions in all three. D, sausage representation of KRASG12D, KRASG12C (-SO2-), and KRASG12C (-S-) structures were extracted from respective MD trajectories and indicate that KRASG12D and KRASG12C (-SO2-) exhibit fluctuations in the KRAS Switch II region, whereas KRASG12C (-S-) exhibits fluctuations in both Switch I and Switch II. E, molecular docking of MRTX-1133, a known G12D inhibitor, to one of the highly populated structural ensembles of KRASG12C (-SO2-) (green) shows similar binding to KRASG12D (pink) as determined by X-ray crystallography (PDB: 7RPZ). The RMSD between MRTX-1133–bound KRASG12D and KRASG12C (-SO2-) is approximately 0.65 Å.