Figure 3.
The HTT allele structure associated with age at HD diagnosis and somatic expansion of the HD allele in blood DNA in African ancestry individuals
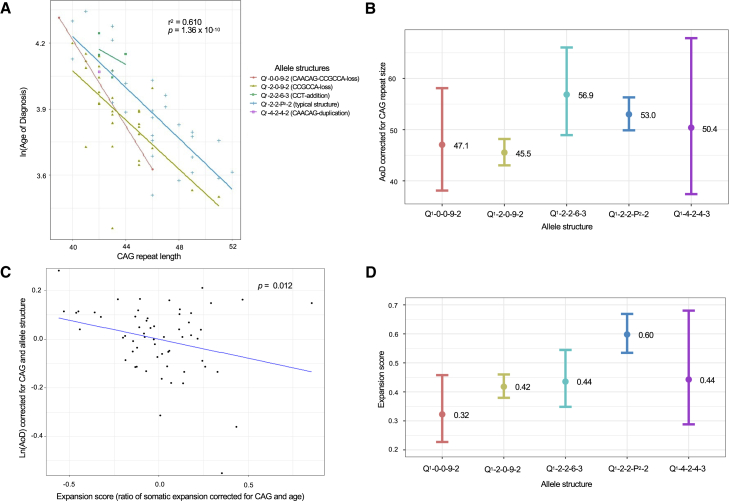
(A) Linear regression analysis testing the association between the log transformed AoD and the inherited CAG repeat length for each disease allele structure revealed a significant association (r2 = 0.61, p = 1.36 × 10−10). The Q1-0-0-9-2 and Q1-2-0-9-2 disease allele structures characterized by the loss of one or more of the intervening sequences had the earliest AoD.
(B) The estimated marginal mean AoD for the disease allele structures, corrected for repeat size. The Q1-2-0-9-2 allele structure had the earliest mean AoD (n = 30, 45.5 years: 95% CI = 43.0–48.2), followed by Q1-0-0-9-2 (n = 2, 47.1 years: 95% CI = 38.1–58.1), Q1-4-2-4-3 (n = 1, 50.4 years: 95% CI = 37.4–67.9), Q1-2-2-P2-2 (n = 31, 53.0 years: 95% CI = 49.9–56.3), and Q1-2-2-6-3 (n = 4, 56.9 years: 95% CI = 48.9–66.0).
(C) Linear regression analysis testing the association between the log transformed AoD (corrected for CAG repeat length and allele structure) and expansion score. Overall, a significant association (p = 0.012) was identified.
(D) The estimated marginal mean expansion score for the allele structures, corrected for CAG repeat length and age at sampling. The Q1-0-0-9-2 (n = 2, 0.32: 95% CI = 0.227–0.458) and Q1-2-0-9-2 (n = 30, 0.42: 95% CI = 0.380–0.460) allele structures had the lowest mean expansion score followed by Q1-2-2-6-3 (n = 4, 0.44: 95% CI = 0.348–0.545), Q1-4-2-4-3 (n = 1, 0.44: 95% CI = 0.288–0.680), and Q1-2-2-P2-2 (n = 31, 0.60: 95% CI = 0.535–0.669).