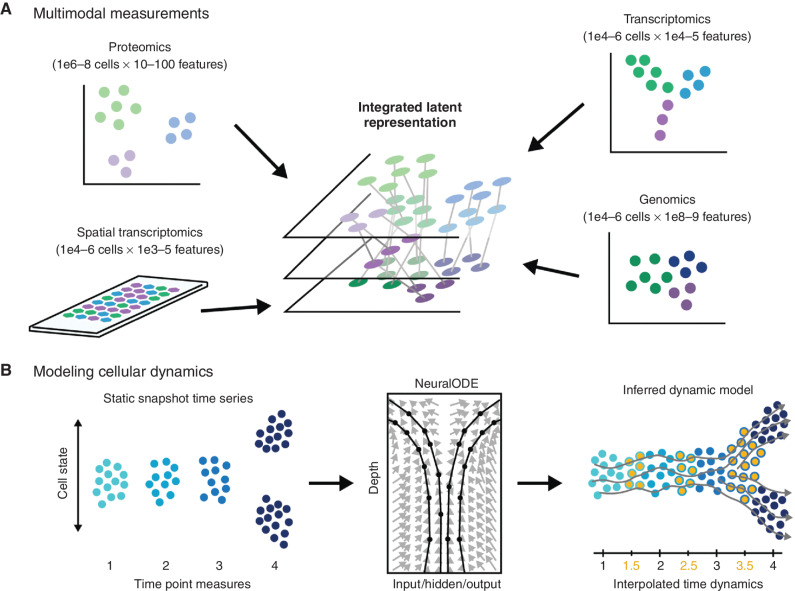
Figure 2.
Modeling temporal dynamics. A, Single-cell populations can be characterized via different omics modalities capturing genomic, transcriptomic, or proteomic information. Resulting data sets may vary significantly in the number of observations and the number of features, and different sets of relationships may exist between the same set of cells depending on which set of features is being examined. Data integration algorithms must be used to merge data sets for joint analysis of multiple data domains. B, In the context of single-cell time-series analyses comprising discrete time point data sets (left), dynamical models based on optimal transport or neural ordinary differential equations (NeuralODE; center) have been used to improve our understanding of biological dynamics by interpolating intervening time point data (orange points, right) to allow inference of dynamic trajectory models (gray lines).