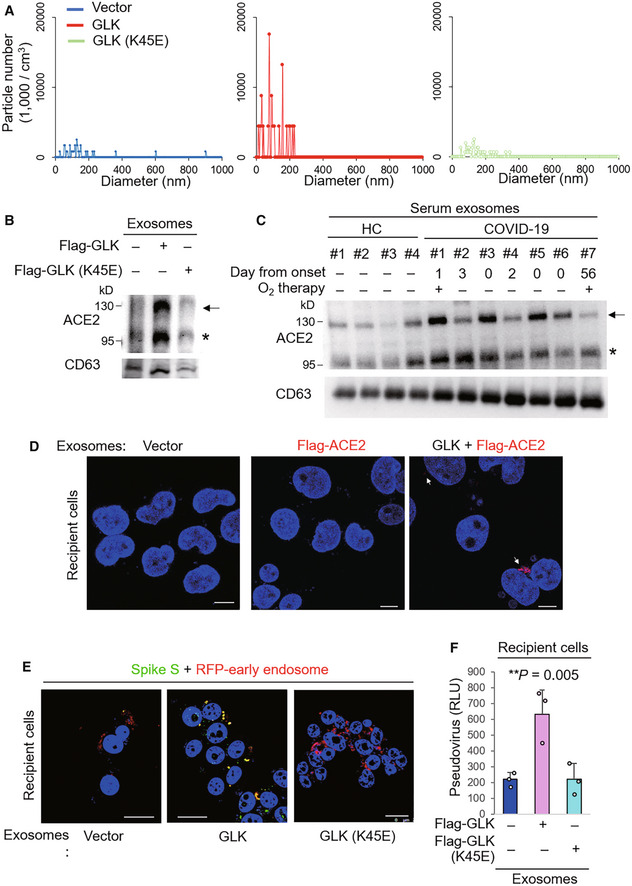
Figure 3. MAP4K3/GLK induces ACE2‐containing exosomes from epithelial cells.
-
AZetaView nanoparticle tracking analysis of particle numbers and sizes of CD63+ extracellular vesicles (EVs) isolated from the supernatants of HCC827 lung epithelial cancer cells, which were transfected with GLK wild‐type or GLK kinase‐dead (K45E) mutant. EVs were isolated sequentially using ExoQuick kits and then ExoQuick ULTRA columns.
-
BImmunoblotting of ACE2 and CD63 proteins in exosomes isolated from GLK wild‐type‐ or GLK (K45E) kinase‐dead‐overexpressing HCC827 cells. Exosomes were isolated sequentially using ExoQuick kits and then ExoQuick ULTRA columns. Arrowhead denotes glycosylated ACE2 proteins; asterisk denotes nonglycosylated ACE2 proteins.
-
CImmunoblotting of ACE2 and CD63 proteins in serum exosomes isolated from the sera of four healthy controls (HC) and seven COVID‐19 patients from NHRI Biobank for COVID‐19 patients in Taiwan (Cohort #3). Serum collection days from the onset are shown; treatment of oxygen therapy on the patient is also indicated. Exosomes were isolated sequentially using ExoQuick kits and then ExoQuick ULTRA columns. Arrowhead denotes glycosylated ACE2 proteins; asterisk denotes nonglycosylated ACE2 proteins.
-
DConfocal microscopy analysis of Flag‐tagged ACE2 proteins (red) in recipient cells after incubation with exosomes for 72 h. Exosomes were isolated sequentially from the supernatants of Flag‐ACE2‐overexpressing or GFP‐GLK plus Flag‐ACE2‐coexpressing HCC827 cells using ExoQuick kits and then ExoQuick ULTRA columns. Arrows denote Flag‐tagged ACE2 proteins. Original magnification, ×630; scale bars, 10 μm.
-
EConfocal microscopy analysis of SARS‐CoV‐2 spike (S) protein (green) and RFP‐labeled early endosomes (red) in exosome‐recipient HCC827 epithelial cells. After incubation with exosomes for 72 h, recipient cells were treated with the spike protein (S) for another 24 h. Yellow color suggests the localization of S protein in early endosomes. Original magnification, ×630; scale bars, 25 μm.
-
FCell entry efficiencies of SARS‐CoV‐2 pseudovirus into exosome‐recipient HCC827 epithelial cells were measured by luciferase activity and presented as relative light units (RLU) at 24 h postinfection. n = 3 (biological replicates).
Data information: In (F), data are presented as means ± SEM. **P‐value < 0.01 (ANOVA test). Data shown are representative results of two (A) or three (B, D, E, F) independent experiments.
Source data are available online for this figure.
