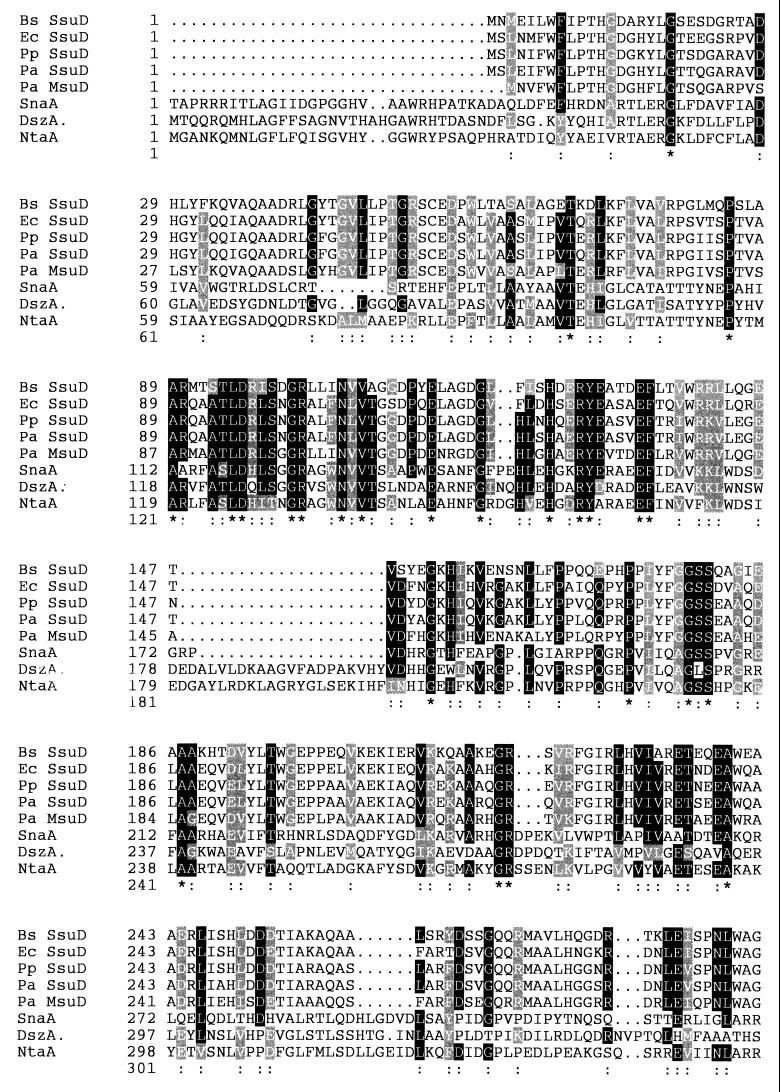
FIG. 2.
Sequence comparison of FMNH2-dependent monooxygenases. Sequence alignment was done with the program CLUSTALW. Amino acid identities (*) and similarities (:) are shown. The proteins are B. subtilis (Bs) SsuD (OrfM) (accession no. L16808), E. coli (Ec) SsuD (Ssi6) (accession no. P80645), P. putida (Pp) SsuD (accession no. AF075709), P. aeruginosa (Pa) SsuD (preliminary results from the Pseudomonas sequencing project [34]), P. aeruginosa MsuD (accession no. AF026067), Streptomyces pristinaespiralis pristinamycin synthase A (SnaA) (accession no. P54991), R. erythropolis DBT oxide monooxygenase (DszA [SoxA]) (accession no. P54995), and Chelatobacter heintzii nitrilotriacetate monooxygenase (NtaA) (accession no. P54989).