Abstract
This study explored the prognostic and therapeutic potentials of multiple Proteasome 26S Subunit, ATPase (PSMC) family of genes (PSMC1-5) in lung adenocarcinoma (LUAD) diagnosis and treatment. All the PSMCs were found to be differentially expressed (upregulated) at the mRNA and protein levels in LUAD tissues. The promoter and multiple coding regions of PSMCs were reported to be differentially and distinctly methylated, which may serve in the methylation-sensitive diagnosis of LUAD patients. Multiple somatic mutations (alteration frequency: 0.6–2%) were observed along the PSMC coding regions in LUAD tissues that could assist in the high-throughput screening of LUAD patients. A significant association between the PSMC overexpression and LUAD patients’ poor overall and relapse-free survival (p < 0.05; HR: >1.3) and individual cancer stages (p < 0.001) was discovered, which justifies PSMCs as the ideal targets for LUAD diagnosis. Multiple immune cells and modulators (i.e., CD274 and IDO1) were found to be associated with the expression levels of PSMCs in LUAD tissues that could aid in formulating PSMC-based diagnostic measures and therapeutic interventions for LUAD. Functional enrichment analysis of neighbor genes of PSMCs in LUAD tissues revealed different genes (i.e., SLIRP, PSMA2, and NUDSF3) previously known to be involved in oncogenic processes and metastasis are co-expressed with PSMCs, which could also be investigated further. Overall, this study recommends that PSMCs and their transcriptional and translational products are potential candidates for LUAD diagnostic and therapeutic measure discovery.
Keywords: biomarker, diagnostic, lung cancer, PSMCs, therapeutic
Introduction
Lung adenocarcinoma (LUAD), which develops along the outer edge of the lungs within glandular cells in the small airways and falls under the umbrella of non-small cell lung cancer (NSCLC), is the most common type of histology, accounting for about 40% of all lung malignancies (Senosain and Massion, 2020; Zheng et al., 2020). Worldwide research on 185 countries suggests that about 11.4% (more than 2.2 million) new cases of lung cancer were diagnosed in 2020, with an almost 18% mortality rate (1.8 million deaths) (Sung et al., 2021). The low survival rate of patients with LUAD can be attributed to the lack of understanding of lung cancer biology, genomics, and host factors that drive the progression of preinvasive lesions, heterogeneity of disease, and patients’ outcomes. Although available diagnosis methods and treatment options have led to the overall decline in the mortality rate from this prevalent cancer, the 5-years survival rate remains below 20% (Hirsch et al., 2017; Myers and Wallen, 2022). Therefore, there is an increasing demand to secure an efficient diagnostic and therapeutic target for LUAD diagnosis and treatment that can significantly aid in the early-stage diagnosis, proper tracking of the patients throughout the cancer stages, and appropriate therapeutic interventions ultimately reducing the medical burden. Investigation of specific prognostic and therapeutic markers for disease stages or tumor types can help develop better screening strategies, improve patients’ prognoses, and assuage the financial burden of the disease (Hirsch et al., 2017; Oberndorfer and Müllauer, 2018; Devarakonda and Govindan, 2019; Zheng et al., 2020). Additionally, exploring molecular features, i.e., genetic variation, aberrant methylation, and immunophenotypes of specific targets can further increase the precision of cancer diagnosis and treatment.
The multiple Proteasome 26S Subunit, ATPase (PSMC) family of genes are reported to be involved in protein degradation, which plays a vital role in regulating the 26S proteasome (Kao et al., 2021). This family of genes is composed of six members, namely, PSMC1, PSMC2, PSMC3, PSMC4, PSMC5, and PSMC6 (PSMC1-6) (Table 1). They partially constitute the formation of the 19S proteasome complex comprised of 19 essential subunits (Gu and Enenkel, 2014). This regulatory complex, in turn, catalyzes the unfolding and translocation of substrates into the 20S proteasome (Kao et al., 2021). Proteasomes control normal cellular function and maintain homeostasis by regulating the optimum degradation of different cellular proteins. However, the upregulated proteasome activity can greatly alter a broad range of crucial cellular processes i.e., DNA replication, transcription, cell cycle and apoptosis (Manasanch and Orlowski, 2017). Given its pivotal roles in the aberrant degradation of the mediators (i.e., activators and inhibitors) of cell cycle and apoptosis regulators upon overproduction in cancer cells, inhibition of the proteasome activity remains a promising target for anticancer therapy development (Sterz et al., 2008; Park et al., 2018).
TABLE 1.
Genomic characteristics of all the subunits in the PSMC gene family.
| Gene | Length of the nucleotide (bp) | Chromosomal location | Length of amino acids | Protein encoded | References |
|---|---|---|---|---|---|
| PSMC1 | 18,903 | 14q32.11 | 440 | 26S protease regulatory subunit 4 (26S proteasome AAA-ATPase subunit Rpt2) | (Tanahashi et al., 1998; National Center for Biotechnology Information (NCBI), 2022) |
| PSMC2 | 41,777 | 7q22.1-q22.3 | 433 | 26S protease regulatory subunit 7 (26S proteasome AAA-ATPase subunit Rpt1) | (Tanahashi et al., 1998; National Center for Biotechnology Information (NCBI), 2022; Shibuya et al., 1992) |
| PSMC3 | 7,705 | 11p11.2 | 439 | 26S protease regulatory subunit 6A (26S proteasome AAA-ATPase subunit Rpt5) | (Hoyle et al., 1997; Tanahashi et al., 1998) |
| PSMC4 | 10,615 | 19q13.11-q13.13 | 418 | 26S protease regulatory subunit 6B (26S proteasome AAA-ATPase subunit Rpt3) | (Tanahashi et al., 1998; National Center for Biotechnology Information (NCBI), 2022; Choi et al., 1996) |
| PSMC5 | 4,875 | 17q23.3 | 406 | 26S protease regulatory subunit 8 (26S proteasome AAA-ATPase subunit Rpt6) | (Tanahashi et al., 1998; National Center for Biotechnology Information (NCBI), 2022; Hoyle et al., 1997) |
| PSMC6 | 21,428 | 14q22.1 | 389 | 26S protease regulatory subunit S10B (26S proteasome AAA-ATPase subunit Rpt4) | (Tanahashi et al., 1998; National Center for Biotechnology Information (NCBI), 2022; Fujiwara et al., 1996) |
As of now, multiple PSMC family genes have been studied in the context of different human diseases including carcinoma. For example, a previous study showed that PSMC6 promotes osteoblast apoptosis and cancer cell proliferation by inhibiting the activation of the PI3K/AKT signaling pathway in an animal model of ovariectomy-induced osteoporosis (Zhang et al., 2020). PSMC2 was found to be upregulated in osteosarcoma (Song et al., 2017), prostate cancer (Chen et al., 2021), pancreatic cancer (Qin et al., 2019), glioma (Zheng et al., 2022), oral squamous cell carcinoma (OSCC) (Wang et al., 2022), and hepatocellular carcinoma (HCC) (Ding et al., 2019; Li et al., 2019; Liu et al., 2021). Moreover, PSMC2 was also reported to promote proliferation and inhibit apoptosis of glioma cells, and its knockdown halted the development and metastasis of prostate cancer (Chen et al., 2021) and progression of OSCC cells by promoting apoptosis via PI3K/Akt pathway and increasing the expression of pro-apoptotic proteins (Wang et al., 2022). PSMC5 is involved in the ubiquitination-dependent degradation of Tln1 and angiogenesis by blocking the miR-214/PTEN/Akt pathway (Li et al., 2019). Knockdown of Proteasome 26S subunit ATPase 3 interacting protein (PSMC3IP) resulted in the suppression of xenograft proliferation and tumorigenesis in the HCC cells (Wang et al., 2022). In a recent study, researchers elucidated the crucial role of PSMC family members and their downstream-regulated genes in breast cancer progression (Kao et al., 2021). However, the collective potential of the PSMC family of genes as candidates to be novel prognostic biomarkers and therapeutic targets in LUAD remains to be unveiled. Out of the six PSMC subunits, a recent systematic study evaluated the differential expression levels and prognostic values of PSMC6 as a high PSMC6 expression was associated with poor prognosis of LUAD, indicating the potential of PSMC6 as a promising therapeutic target for LUAD (Zhang et al., 2021). Though the study mentioned earlier focused on the prognostic power of PSMC6 in LUAD, the molecular characterization, i.e., genetic alteration frequency, aberrant methylation, and immune phenotypes of the PSMC family genes in LUAD, which could further assist in diagnostic and therapeutic development, remains unstudied from a holistic perspective.
This study evaluated the prognostic and therapeutic significance of the multiple Proteasome 26S Subunit, ATPase (PSMC) family of genes (PSMC1-5) in LUAD utilizing a web-based database mining approach. Since the prognostic value of PSMC6 has been studied in the context of LUAD, this member of PSMC family was not considered in this study (Zhang et al., 2021). Using a bioinformatics approach, we attempted to determine the expression patterns, methylation patterns, mutations, and copy number alterations of the PSMC genes in LUAD tissues (Figure 1). Furthermore, we examined the correlation between PSMC overexpression and the clinical features and different survival rate of LUAD patients. We also assessed the association between PSMC expression and abundance of tumor-infiltrating immune cells and co-expressed genes of PSMCs and their functional enrichment in LUAD patients. Our study should contribute to understanding the predictive roles of PSMCs and their transcriptional and translational products in LUAD development, progression, and prognosis, which should help further research work and clinical development of PSMC-based diagnostics and therapeutics for LUAD.
FIGURE 1.

Strategies utilized in the database mining approach employed in the overall study.
Materials and Methods
Expression Analysis of PSMC genes in Normal Lung and LUAD Tissues
PSMC gene expression pattern at the mRNA level in normal lung and cancerous LUAD tissues was determined using the OncoDB server (http://oncodb.org/, accessed on: 7 April 2022). OncoDB is an online platform that allows users to explore the differential gene expression (DGE) pattern in normal and corresponding cancerous tissues from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases (Su et al., 2007). The differential expression pattern of PSMCs was evaluated between the log2 TPM (transcript per million) normalized RNA sequencing data of LUAD and adjacent normal lung tissue samples in the OncoDB server. The result was then analyzed based on the log2 fold change (log2FC) and false discovery rate adjusted p-value cutoff of the DGE analysis. After that, the Expression Atlas (https://www.ebi.ac.uk/gxa/home, accessed on: 7 April 2022) web-based tool was utilized to discover the mRNA level expression pattern of the PSMCs in a total of 68 different types of LUAD cell lines (Papatheodorou et al., 2018). Finally, the Human Protein Atlas (HPA) (https://www.proteinatlas.org/, accessed on: 7 April 2022) server was used to determine the protein level expression of PSMCs in normal lung and LUAD tissues by analyzing the immunohistochemistry (IHC) images (at 200 µm length) of the LUAD samples and adjacent normal lung tissues (Pontén et al., 2008). The Pathology and Tissues modules of the HPA server were explored to optimize the differences in PSMC protein expression between normal and cancerous lung tissues.
Analysis of the Promoter and Coding Sequence Methylation of PSMC Genes in LUAD Patients
The UALCAN server (http://ualcan.path.uab.edu/, accessed on: 7 April 2022) was utilized to examine the promoter methylation pattern of PSMC genes in LUAD tissues (Chandrashekar et al., 2017). TCGA database (integrated with UALCAN server) was selected as the basis set for the experiment. The analysis was carried out by performing a student’s t-test between the methylation data of the test (LUAD samples) and control (adjacent normal lung tissue samples) variables. Finally, the result was checked and validated based on a significant p-value cutoff of <0.05. Next, the methylation pattern of the DNA sequence of PSMC coding genes was studied using the UCSC Xena browser (https://xenabrowser.net/, accessed on: 7 April 2022) (Goldman et al., 2019). In this step, the integrated TCGA LUAD samples (n = 706) were again selected to observe the PSMC coding sequence methylation using the methylation 450k array data. The samples for which methylation data were not available were omitted during the analysis. Finally, the GSCA server was used to confirm the association between PSMC methylation and gene expression in LUAD tissues (http://bioinfo.life.hust.edu.cn/GSCA/#/, accessed on: 7 April 2022) (Liu et al., 2018). The impact of PSMC methylation on the survival rate of LUAD patients was also evaluated from the GSCA tool.
Examination of Mutation and Copy Number Alteration in PSMC Genes Across Different LUAD Studies
The cBioPortal server (https://www.cbioportal.org/, accessed on: 7 April 2022) was accessed to analyze the mutation and copy number alteration (CNA) in PSMC genes across a wide number LUAD study samples (Gao et al., 2013). The data deposited by MSKCC, Broad, OncoSG, TDP, CPTAC, and others including more than 2,598 patients’ samples over nine studies were searched for PSMC mutation and CNA analysis in LUAD patients. The OncoPrint summary of the overall mutations of the selected PSMCs in different LUAD studies was inspected. Next, the bar diagram representing the type of genetic alterations in PSMC coding genes was also analyzed. The relation between the overall survival (OS) of LUAD patients and PSMC gene alteration was evaluated from this server. The parameter values were kept at defaults during the analysis in cBioPortal server. Finally, the correlation between the CNAs present in PSMCs their mRNA level expression in LUAD tissues was discovered in the form of a bubble plot using the mutation module in GSCA (accessed on: 7 April 2022) server.
Analysis of the Correlation Between the PSMC Overexpression and Clinical Features of LUAD Patients
The association between the PSMC gene overexpression and LUAD patients’ clinical features and demographic status, i.e., age, individual cancer stages, and nodal metastasis status, was evaluated from the UALCAN server (accessed on: 7 April 2022). UALCAN is a comprehensive, user-friendly online tool that enables users to access omics data in cancer biomarker discovery and target validation. The TCGA LUAD samples were selected for the association analysis with our genes of interest in this study. The analysis result was considered significant based on the p-value cutoff of <0.05 found in the student’s t-test, and the expression profile was retrieved as box plots with transcript per million (TPM) reads unit.
Assessing the Association of the PSMC Expression with LUAD Patients’ Survival Rate
The association between OS of LUAD patients and PSMC expression was established using the GEPIA 2 server (http://gepia2.cancer-pku.cn/, accessed on: 7 April 2022) (Tang et al., 2019). GEPIA 2 involves 9,736 tumors and 8,587 normal samples from GTEx and TCGA projects of RNA sequencing data, and this tool facilitates different transcriptional analyses, i.e., the analysis of correlation and differential expression across different normal and tumor tissues. Finally, the relation between the PSMC expression and LUAD patients’ relapse-free survival (RFS) was also determined from the GEPIA 2 (accessed on: 29 April 2022) server. The result of the experiment was then analyzed based on the p-value and hazard ratio (HR) of LUAD patients in relation to the differential level of PSMC expression represented in the Kaplan-Meier (KM) plot of survival analysis. The parameter values were kept at default during the analysis in GEPIA 2 server.
Analysis of Correlation Between the Abundance of Tumor-Infiltrating Immune Cells and the PSMC Expression in LUAD Patients
The association between abundance of immune cells and PSMC expression in LUAD patients was determined utilizing the immune module of the GSCA database (accessed on: 7 April 2022). GSCA is a highly inclusive database that helps analyze different genomic association features and cancer patients’ clinical outcomes across different forms of cancer. Moreover, it also aids in the analysis of the correlation between different gene expressions, gene mutations, and the expression level of 24 different types of immune cells in different cancer patients. Every selected PSMC gene was queried against the abundance of immune cells like B Cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, natural killer (NK) cells in LUAD microenvironment. Finally, the association between the PSMC expression and the abundance of different immunomodulators in LUAD patients was determined from the TISIDB server (http://cis.hku.hk/TISIDB/, accessed on: 7 April 2022) (Ru et al., 2019). The result of immune cell and modulator’s infiltration level was analyzed based on p-value and correlation coefficient.
Identification of the Co-Expressed Genes of PSMCs in LUAD Patients and Their Functional Enrichment Analysis
The co-expressed genes of PSMCs were identified using the TCGA LUAD database (Firehose, Legacy) from the cBioPortal server (accessed on: 7 April 2022). After that, the top 300 positively co-expressed genes of each PSMC were selected based on p-value and correlation coefficient, which were then used to identify the overlapping neighbor genes utilizing the InteractiVenn online tool (http://www.interactivenn.net/, accessed on: 7 April 2022) (Heberle et al., 2015). The overlapping neighbor genes of PSMCs in LUAD tissues were then used in gene ontology terms, i.e., biological processes (BP), molecular function (MF), cellular component (CC), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis from the Enrichr server (https://maayanlab.cloud/Enrichr/, accessed on: 7 April 2022) (Kuleshov et al., 2016). The result of the functional enrichment analysis was then visualized and retrieved in the form bubble plot using the ImageGP online and publicly available tool (http://www.ehbio.com/ImageGP/, 7 April 2022) (Chen et al., 2022).
Validation of the PSMC Gene Expression and its Correlation with LUAD Patients’ Survival Rate in the Public Dataset
In this step, we evaluated the pattern of PSMC mRNA expression in two independent microarray datasets, i.e., GSE1037 and GSE116959 from National Center for Biotechnology Information-Gene Expression Omnibus database (Barrett et al., 2012). GSE1037 contains total mRNA expression profiles of 105 lung cancer and adjacent normal lung tissue samples out of which 12 and 19 samples correspond to LUAD and adjacent normal tissues, respectively, which were utilized in our analysis (Jones et al., 2004). On the other hand, GSE116959 contains mRNA expression profiles of 57 LUAD and 11 adjacent normal lung tissue samples (Moreno Leon et al., 2019). Data normalization, log2 transformation, and expression value were calculated on the selected datasets using the BioConductor package in R studio (Gentleman et al., 2004; Allaire, 2012). The expression pattern was then visualized in the form of boxplot using ggplot2 package (Wickham et al., 2016). Moreover, we also examined the correlation between PSMC1-5 expression and LUAD patients' OS in GSE31210 microarray dataset that contains the clinical and mRNA expression profile of 226 LUAD patients (Okayama et al., 2012). A log-rank t test was applied between the higher and lower PSMC expressing LUAD patients using the survival and survminer packages in R studio and the result was retrieved in the form of KM plot (Kassambara et al., 2017).
Results
mRNA and Protein Level Differential Expression of PSMC Genes in Normal Lung and LUAD Tissues
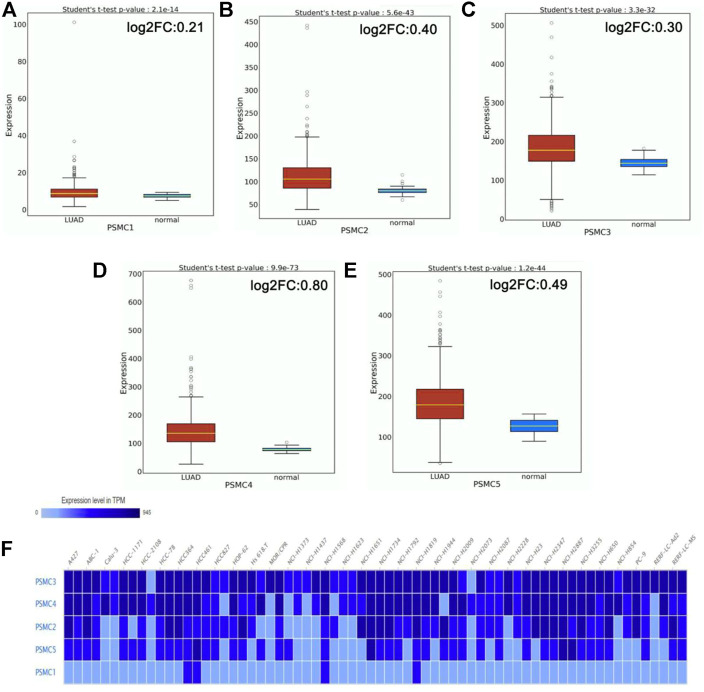
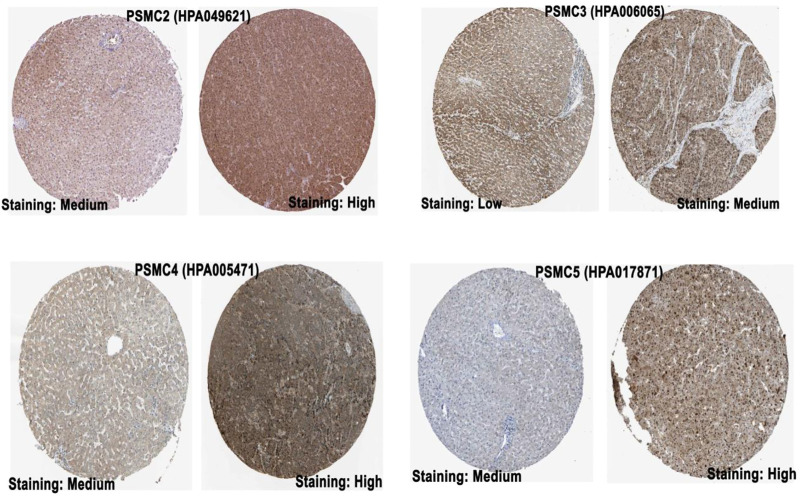
The mRNA level expression of PSMCs in normal lung and LUAD tissues was analyzed from the OncoDB server. All the PSMC genes showed higher expression levels in LUAD tissues than in normal lung tissues (Figure 2). Moreover, PSMC4 (log2FC: 0.80) showed the highest difference of expression between the test and control group followed by PSMC5 (log2FC: 0.49), PSMC2 (log2FC: 0.40), PSMC3 (log2FC: 0.30) and PSMC1 (log2FC: 0.21). Thereafter, the expression pattern of our genes of interest was observed across 68 different LUAD cell lines and PSMC3 was discovered to be overexpressed inmost of the selected cell lines followed by PSMC4 and PSMC2 (Figure 2F). On the contrary, as in par with the previous result, PSMC1 showed the least overexpression in all the LUAD cell lines. Overall, all the selected PSMCs showed higher expression levels in HCC461 and NCI-H1819 cell lines. Thereafter, the protein level expression pattern of the PSMCs in LUAD and their corresponding normal tissues was analyzed from the HPA server. PSMC2 showed medium staining against the administered antibody (HPA049621) in normal lung tissues, whereas a stronger staining was recorded in the LUAD tissues (Figure 3). PSMC3 demonstrated a low staining pattern in the normal lung tissues and medium staining in the LUAD tissues. Moreover, both PSMC4 and PSMC5 exhibited medium staining in the normal lung tissues, whereas a high level of staining was observed in the LUAD tissues.
FIGURE 2.
mRNA level differential expression patterns of PSMC1 (A), PSMC2 (B), PSMC3 (C), PSMC4 (D), and PSMC5 (E) in normal lung and LUAD tissues observed from the OncoDB server. The expression values are presented in the TPM unit (log2 transformed). The red colored box represents LUAD samples, and the green colored box represents normal samples. The expression of the PSMCs in different LUAD cell lines (F). The color gradient represents the expression value of PSMCs in TPM units in different cell lines, i.e., low intensity corresponds to a lower TPM and high intensity corresponds to a higher TPM, while the TPM value escalates with an increasing gradient from low to high. FC: fold change.
FIGURE 3.
IHC images (visualized at 200 µm) delineating the protein level expression of PSMCs in normal lung (left) and LUAD tissue (right) from the HPA server. The name of the corresponding antibody used for IHC staining has been indicated inside the parentheses in addition to the gene name. The representative image for PSMC1 was not found (Source: The Human Protein Atlas; https://www.proteinatlas.org/).
Promoter and Coding Sequence Methylation Status of PSMC Coding Genes in LUAD Tissues
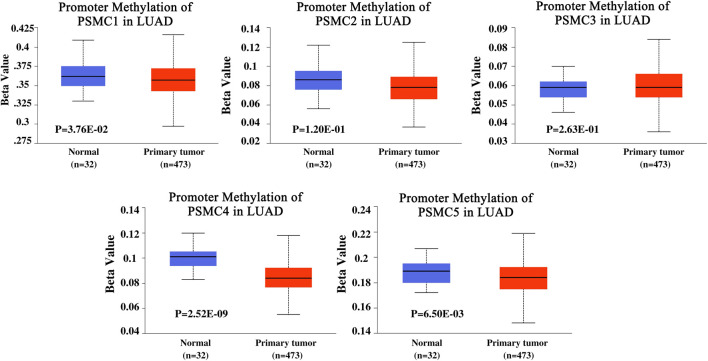
The promoter methylation pattern of the PSMC genes in LUAD and normal lung tissues was examined from the UALCAN server. PSMC1 gene coding promoter in LUAD tissues was found to be less methylated than in the normal lung tissues (p = 3.76e-02) (Figure 4). Although the PSMC2 and PSMC3 promoters were observed to be less methylated in LUAD tissues, the association was not significant (p > 0.05). Additionally, PSMC4 (p = 2.52e-09) and PSMC5 (p = 6.50e-03) promoters were also found to be less methylated in LUAD tissues compared to the normal lung tissues. The coding sequence methylation analysis of the PSMCs in LUAD tissues from the UCSC Xena browser revealed that the selected PSMCs might have distinct coding sequence methylation patterns. For example, PSMC2, PSMC3, and PSMC4 signified that their coding regions might have the mostly methylated regions at the 3’ end of the sequence as indicated by the elevated beta value in red-colored regions (Supplementary Figure S1). On the contrary, PSMC1 showed a completely different pattern of methylation in which most of the CpG islands might cover the 5’ end and a slight upstream region from the 3’ end of the coding sequence. In the case of the PSMC5 methylation pattern, the red landscapes at the 5’ end indicated that the initial region of the coding sequence might be hypermethylated. Finally, the effect of methylation in PSMC genes on their mRNA expression level specific to LUAD tissues was determined from the GSCA server. Unsurprisingly, methylation was negatively correlated with the PSMC1, PSMC2, PSMC4, and PSMC5 mRNA expression in LUAD tissues (Supplementary Figure S2). Additionally, PSMC5 hypomethylation was found to be associated with poor OS (p = 0.043) and progression-free survival (PFS) (p = 0.017) in LUAD patients (Supplementary Figure S3).
FIGURE 4.
Promoter methylation pattern of PSMC genes in LUAD and normal lung tissues. Significant and distinct differential methylation patterns of different PSMCs in LUAD tissues was observed compared to the normal lung tissues. Normal: samples were collected from normal tissues adjacent to the cancerous tissues of LUAD patients within TCGA cohorts without any demographic and clinical stratification (Source: The Cancer Genome Atlas). Beta value cut-offs in the range of 0.7 to 0.5 indicate hypermethylation; 0.3–0.25 indicates hypomethylation.
Frequency of PSMC Mutation and Copy Number Alterations in LUAD Tissues
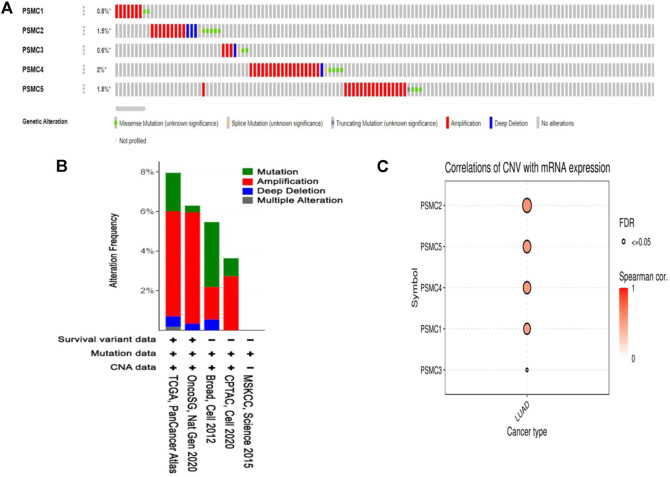
The mutation and copy number alteration frequency of PSMC genes across different LUAD studies were evaluated from the cBioPortal online tool. PSMC1, PSMC2, PSMC3, PSMC4, and PSMC5 showed an alteration frequency of 0.8, 1.5, 0.6, 2, and 1.8%, respectively, across different LUAD studies (Figure 5A). The analysis reported the presence of different detrimental genetic alterations i.e., amplification, deep deletion, and splice sites across the PSMC coding regions in LUAD samples. Moreover, a number of missense mutations was also recorded in the PSMC coding regions carrying the potential to interfere with the protein functions. The copy number alteration (CNA) frequency analysis revealed the shreds of evidence of a large number of amplification events across the selected LUAD studies responsible for the genetic alterations in PSMC genes in LUAD patients (Figure 5B). Thereafter, the association between PSMC CNA in LUAD tissues and their expression patterns was established from the GSCA server. All the PSMCs showed a significant and positive correlation between the number of CNA events and their expression levels (Figure 5C). Finally, the effect of the mutations and CNAs present in PSMC genes on LUAD patients’ OS was also determined from this server. It was observed that PSMC mutations are significantly and negatively correlated to the LUAD patients’ OS (p = 1.19e-04) (Supplementary Figure S4). Altogether, PSMCs altered LUAD patients had a poor OS (median survival ∼49 months) compared to the unaltered patients (median survival ∼66 months).
FIGURE 5.
Mutation analysis report on PSMC genes in LUAD patients presented in OncoPrint diagram (A). The colored region of the column represents different alterations, and the alternation frequency is a fraction of the colored columns over the total columns. Amplification was found to be the most prevalent form of genetic alteration in all PSMC genes across different LUAD studies. The distribution of mutation and CNA in PSMC genes across different LUAD studies is oriented in bar diagram (B). The y-axis represents the percentage of total samples mutated in each LUAD study correspondingly presented in the x-axis. Bubble plot representing the positive correlation between the CNAs present in PSMC genes and their mRNA level expression in LUAD tissues (C).
Association Between the PSMC Overexpression and LUAD Patients’ Clinical Features
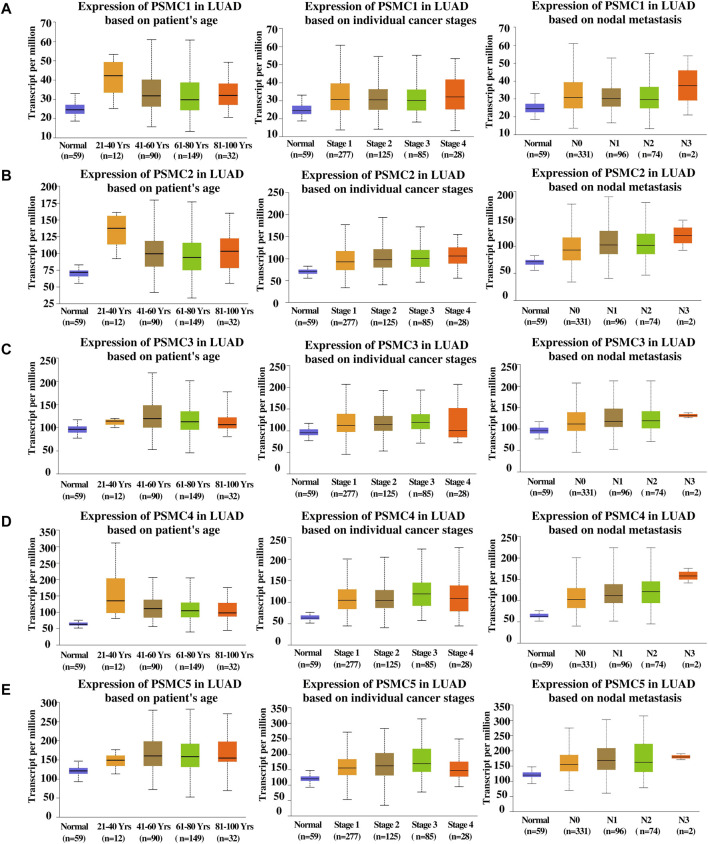
The association between the PSMC overexpression and LUAD patients’ clinical characteristics was determined from the UALCAN server. Although a noticeable rise in the PSMC expression in the 21–40 years age group compared to the normal was found, PSMC1, PSMC3, and PSMC4 genes did not show significant association as observed from student’s t-test performed between normal and cancerous samples (p > 0.05) (Supplementary Table S1) (Figure 6). Moreover, PSMC1-4 showed a marginal reduction in the expression among other age groups except for 21–40 years in LUAD tissues though the level remained above the normal lung tissue expression level whereas PSMC5 showed a significant increase in expression in accordance with advancing age groups (p < 0.05) (Figure 6). On the contrary, in terms of cancer stages, PSMC1 showed the highest expression level in stage 1 and stage 4 whereas the intermediate stages showed a downward trend in expression in LUAD tissues although the expression level still remained above that in normal lung tissues (p < 0.001) (Supplementary Table S1) (Figure 6). In the case of other PSMCs, all the genes showed a marginal increment in their expression levels in accordance with aggressive cancer stages with a slight decline in stage 4 in LUAD tissues (p < 0.05). As in par with the PSMC1 expression level in comparison with cancer stages, its expression level was also found at the highest threshold in N0 and N3 of the nodal metastasis status group in LUAD patients. However, the association between the expression level of PSMC1, PSMC2, and PSMC4 and N3 lymph node metastatic groups was not discovered to be significant (p > 0.05) (Supplementary Table S1) (Figure 6). In contrast, other PSMCs showed significant overexpression in accordance with the advancing metastasis stage in LUAD patients (p < 0.04).
FIGURE 6.
Pattern of PSMC1 (A), PSMC2 (B), PSMC3 (C), PSMC4 (D), and PSMC5 (E) overexpressions in relation to LUAD patients’ age, individual cancer stages, and nodal metastasis status represented in box plots. Several significant associations between PSMCs expression levels and LUAD patients’ clinical features were observed. Normal: samples collected from normal tissues adjacent to the cancerous tissues of LUAD patients within TCGA cohorts without any demographic and clinical stratification (Source: The Cancer Genome Atlas). Please correspond to Supplementary Table S1 for a detailed observation of the clinical parameters.
Relation Between the PSMC Expression and LUAD Patients’ Survival Rate
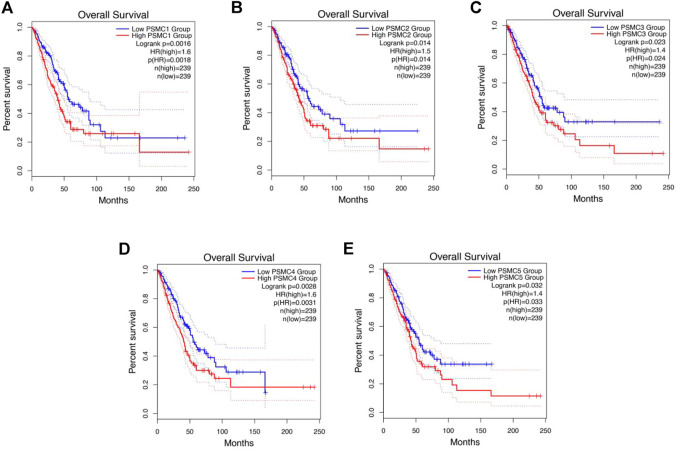
The relation between PSMCs expression and LUAD patients’ OS and RFS was established using the GEPIA 2 server. The report of the survival analysis was retrieved in the form of Kaplan–Meier plot. The analysis revealed that PSMC1 overexpression is negatively correlated with the OS of LUAD patients [p = 0.0016; Hazard Ratio (HR): 1.6] (Figure 7A). Moreover, the PSMC1 overexpression was also responsible for the poor RFS of LUAD patients (p = 0.011; HR: 1.5) (Supplementary Figure S5A). Similarly, PSMC2 overexpression was discovered to be associated with the worsening OS (p = 0.014; HR: 1.5) in LUAD patients (Figure 7B). Remarkably, the PSMC2 overexpression was found to be responsible for poor RFS in LUAD patients as observed by an HR of 1.2 (p = 0.008) (Supplementary Figure S5B). In the case of PSMC3 expression, its overexpression was significantly and negatively correlated to the OS (p = 0.023; HR: 1.4) (Figure 7C). Though the association between PSMC3 and LUAD patients’ RFS was not found to be significant, a noticeably lower p-value of 0.077 and an HR of 1.3 suggested that the overexpression of PSMC3 might be responsible for poor RFS (Supplementary Figure S5C). A significant association was also observed between PSMC4 overexpression and the poor OS of LUAD patients from the report of the survival analysis (p = 0.003; HR: 1.6) (Figure 7D). Again, the expression of PSMC4 was found to be accounted for the worse RFS in LUAD patients also (p = 0.0079; HR: 1.2) (Supplementary Figure S5D). Finally, the PSMC5 expression was also discovered to be negatively associated with the OS (p = 0.03; HR: 1.4) and RFS (HR: 1.3) of LUAD patients, though the association with RFS was not significant (p = 0.089) (Figure 7E) (Supplementary Figure S5E).
FIGURE 7.
Kaplan–Meier plot representation of PSMC1 (A), PSMC2 (B), PSMC3 (C), PSMC4 (D), and PSMC5 (E) expressions and their relation with the OS of LUAD patients. The red color plot represents the high PSMC-expressing LUAD patients, and the blue color plot represents the low PSMC-expressing group. The vertical tick mark within the plot indicates an event (death). A significant negative association was observed between PSMCs expression and LUAD patients’ OS (p < 0.05, HR: <1.4).
Association Between the PSMC Expression and the Abundance of Tumor-Infiltrating Immune Cells and Immune Modulators in LUAD Patients
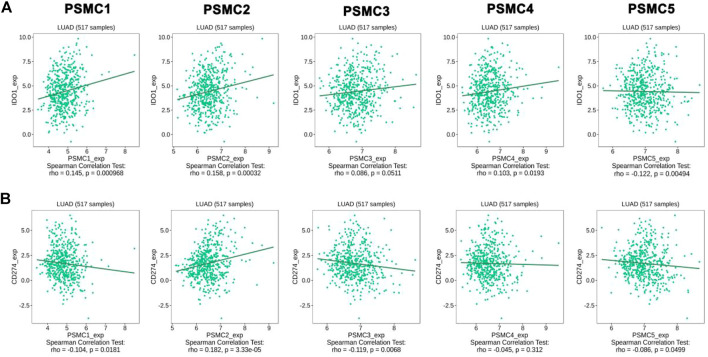
The association between the PSMC expression and the abundance of different immune cells in LUAD tissues was evaluated from the GSCA server. A significant positive correlation between PSMC1 expression and B cell (Cor: 0.09; p = 0.016) and CD8+ T cell (Cor: 0.13; p = 0.009) was observed in LUAD tissues. However, a negative correlation was observed between CD4+ T cell (Cor: −0.27; p = 3.29e-11) and PSMC1 expression (Supplementary Table S2). PSMC2 expression showed significant association with the abundance level of B cell (Cor: 0.16; p = 6.97e-05), CD8+ T cell (Cor: 0.09; p = 0.02) and dendritic cell (DC) (Cor: 0.21; p = 1.13e-07). On the contrary, PSMC2 expression showed significant negative correlation with CD4+ T cell (Cor: -0.36; p = 2.34e-19), Natural Killer (NK) cell (Cor: -0.14; p = 0.0006), and Neutrophil (Cor: -0.12; p = 0.02) abundance level in LUAD and other surrounding tissues (Supplementary Table S2). In case of PSMC3 expression, it showed significant positive association with B cell (Cor: 0.14; p = 0.004), CD8+ T cell (Cor: 0.15; p = 0.002), and Dendritic Cell (DC) (Cor: 0.12; p = 0.003). However, PSMC3 exhibited negative association with CD4+ T cell (Cor: −0.28; p = 7.01E-12), NK cell (Cor: −0.13; p = 0.001) and Neutrophil (Cor: −0.08; p = 0.04) infiltration levels in LUAD patients (Supplementary Table S2). PSMC4 and PSMC5 showed a positive association with B cell, CD8+ T cells, monocytes and DC (Cor: > 0.09, p < 0.05) abundance levels in LUAD microenvironment. A significant negative association between the CD4+ T cell, NK cell, Macrophage and Neutrophil production level and PSMC4 and PSMC5 expression levels was observed (p < 0.05) (Supplementary Table S2). Thereafter, the association between the PSMCs overexpression and different immunoinhibitor infiltration levels in LUAD and adjacent tissues in the microenvironment was established from the TISIDB server. Later, the immunomodulators that showed significant association, i.e., CD274 and IDO1 with the PSMC expression, were inspected. PSMC1, PSMC2, PSMC4, and PSMC5 showed positive correlation with IDO1 expression levels in LUAD tissues (p < 0.05) (Figure 8A). Moreover, the CD274 abundance levels in LUAD tissues showed a negative association with PSMC1, PSMC3, PSMC4, and PSMC5 expression levels in LUAD tissues (p < 0.05) (Figure 8B). Only PSMC2 showed a positive association with CD274 infiltration levels.
FIGURE 8.
Association between the PSMC expression and the infiltration levels of IDO1 (A) and CD274 (B) in LUAD tissues. Significant positive and negative correlations were observed in between PSMC expression levels and immunomodulator expression levels.
Co-Expressed Genes of PSMCs in LUAD Tissues and Their Functional Enrichment Analysis
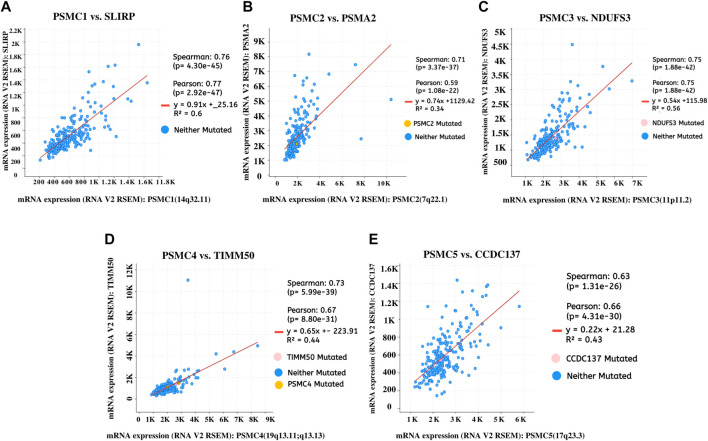
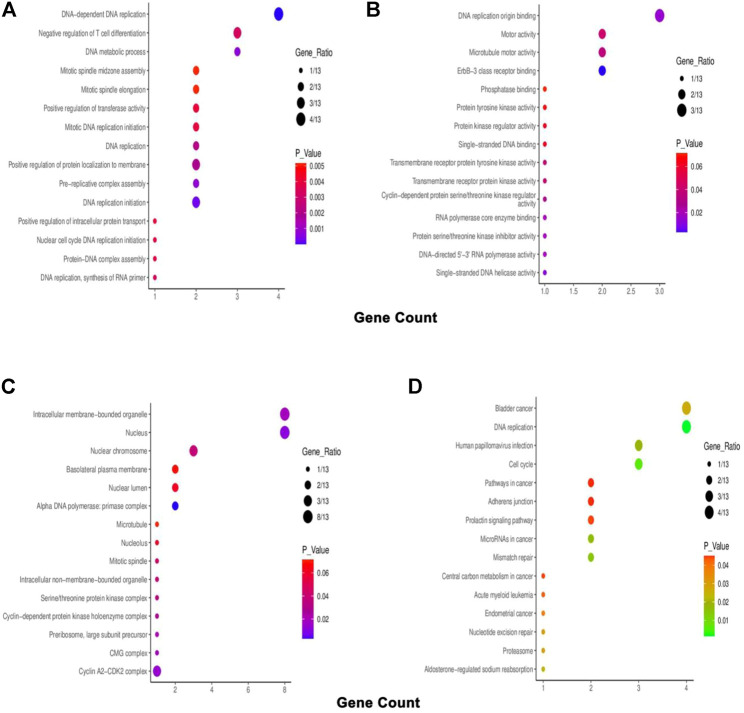
The top co-expressed gene of each PSMC in LUAD samples was identified from the cBioPortal server. PSMC1 was discovered to be highly co-expressed with the SRA stem-loop interacting RNA-binding protein (SLIRP) coding gene in LUAD tissues (Cor: 0.76, p = 4.30e-45) (Figure 9A). Proteasome 20S subunit alpha 2 (PSMA2) gene showed the highest co-expression association with the PSMC2 gene (Cor: 0.76, p = 4.30e-45) (Figure 9B). Moreover, the PSMC3 gene was found to be most highly co-expressed with the NADH: Ubiquinone Oxidoreductase Core Subunit S3 (NDUFS3) gene in LUAD tissue samples (Cor: 0.75; p = 1.88e-42) (Figure 9C). Lastly, PSMC4 and PSMC5 genes were observed to have the highest level of co-expression with Translocase of Inner Mitochondrial Membrane 50 (TIMM50) (Cor: 0.75; p = 5.99e-39) and Coiled-coil Domain Containing 137 (CCDC137) genes (Cor: 0.63, p = 1.31e-26), respectively (Figures 9D,E). The analysis report also suggested the presence of mutated copies of both the PSMC genes and co-expressed genes within the samples except for PSMC1. Thereafter, the overlapping neighbor genes from the top 300 positively co-expressed genes of each PSMC family member in LUAD tissues were identified using the Venn diagram (Supplementary Figure S6). The analysis revealed 13 genes, i.e., PSMB3, MRTO4, RFC2, TACO1, PRIM1, MCM3, KIF23, CCNA2, ERBB2, IRF1, PDCD45, SFN, and TOX3, which are overlapped among the top 300 positively co-expressed genes of PSMCs. Afterward, the overlapping neighbor genes were investigated to understand their differential expression pattern in LUAD tissues. All the genes (except for PDCD45) showed significant overexpression in LUAD tissues compared to the normal lung tissues and only IRF1 showed under-expression (Supplementary Figure S7). Finally, the overlapping genes were used in the functional enrichment analysis delineating different gene ontology terms, i.e., biological processes, molecular functions and cellular components, and the KEGG pathway. The biological process analysis revealed that the highest ratio of the genes is involved in DNA replication, negative regulation of T-cell differentiation, DNA metabolic processes, and mitotic spindle assembly (Figure 10A). The major molecular functions of the queried genes were DNA replication origin binding, phosphatase binding, motor activity, and microtubule motor activity (Figure 10B). The overlapping genes were predominantly operating in intracellular membrane-bound organelles, nucleus, and basolateral plasma membrane as observed from the cellular component analysis (Figure 10C). The KEGG pathway analysis on the overlapping neighbor genes of PSMCs in LUAD tissues reported that most of the genes are involved in pathways associated with bladder cancer, DNA replication, cell cycle, human papillomavirus infection, and so forth (Figure 10D).
FIGURE 9.
Top positively co-expressed genes of PSMC1 (A), PSMC2 (B), PSMC3 (C), PSMC4 (D), and PSMC5 (E) in LUAD tissues obtained from the TCGA LUAD study (Firehose Legacy) through the cBioPortal server. The expression values were compared in the form RNA-seq V2 RSEM normalized scores of the sequencing reads. RSEM: RNA-seq by expectation maximization.
FIGURE 10.
Bubble plots representing the enriched gene ontology terms of the overlapping 13 neighbor genes of PSMCs in LUAD tissues: (A) biological processes, (B) molecular function, (C) cellular component, and (D) KEGG pathway.
Validation of the Differential Expression and Clinical Relevance of PSMC Genes in LUAD Tissues from the Public Dataset
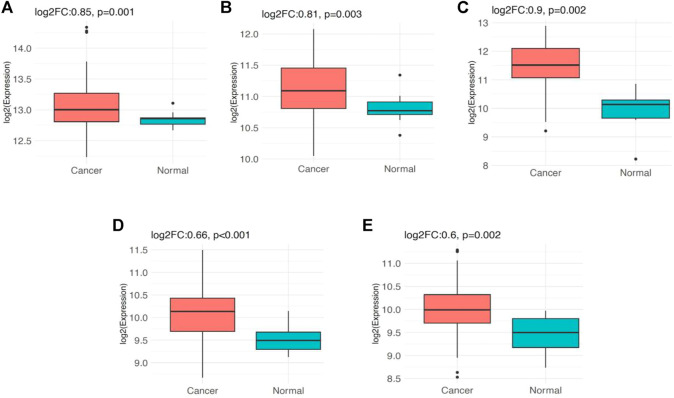
The analysis of PSMC gene expression in GSE116959 dataset revealed that PSMC1 gene is significantly overexpressed (log2FC: 0.85; p = 0.001) in LUAD tissues (n = 57) compared to the adjacent normal lung tissues (n = 11). Similarly, PSMC2 (log2FC: 0.81; p = 0.003), PSMC3 (log2FC: 0.9; p = 0.002), PSMC4 (log2FC: 0.66; p < 0.001) and PSMC5 (log2FC: 0.6; p = 0.002) also showed overexpression in LUAD tissues compared to the normal tissues (Figure 11). Additionally, the expression analysis of PSMC genes in GSE1037 dataset also reported that all the PSMCs (PSMC1-5) are significantly overexpressed (log2FC: >0.66; p < 0.05) in LUAD tissues (n = 12) than in normal lung tissues (n = 19) (Supplementary Figure S8). Thereafter, we examined the association between the PSMC expression and LUAD patients’ OS from GSE31210 (n = 226) dataset by performing a log-rank t-test between the higher and lower PSMC-expressing patients. Though the overexpression of PSMC1 was found to be negatively associated with the OS of LUAD patients, the correlation was not discovered to be significant. On the contrary, PSMC2-5 overexpression was reported to be significantly and negatively associated with the OS of LUAD patients (p < 0.05) (Supplementary Figure S9).
FIGURE 11.
Expression pattern of the PSMC genes in LUAD tissues (n = 57) and adjacent normal lung tissues (n = 11) obtained from the GSE116959 dataset: (A) PSMC1, (B) PSMC2, (C) PSMC3, (D) PSMC4, and (E) PSMC5. All the PSMCs were found to be significantly overexpressed in LUAD tissues compared to the adjacent normal lung tissues (log2FC: >0.59, p < 0.05)
Discussion
This study explored the prognostic values of the PSMC family of gene expression in LUAD, taking advantage of the database mining approach. Initially, the differential expression pattern of all the selected PSMCs in LUAD and its corresponding adjacent normal lung tissues was evaluated. Given that cancer development is a multistep process controlled by various biological processes, differential gene expression analysis allows the understanding of the possible involvements of particular genes in the oncogenic development of a healthy cell (Bashyam, 2002; Liang and Pardee, 2003; Kim et al., 2015). This study found that all the PSMCs are highly expressed in LUAD tissues compared to the normal lung tissues both at the mRNA and protein level, suggesting their possible functions in LUAD development and progression (Figure 2 and Figure 3). Although the expression pattern of the selected genes (PSMC1-5) in LUAD tissues remains unstudied to date, previously PSMC6 was found to be overexpressed at both mRNA and protein levels in LUAD tissues than in adjacent normal lung tissues (Zhang et al., 2021). Moreover, all the genes of our interest also showed higher expression levels in different LUAD cell lines.
DNA methylation is one of the epigenetic drivers in cancer development and progression. Usually, promoter methylation regulates the gene activation and silencing and aberrant methylation is commonly associated with the up or downregulation of different genes in cancer cells (Liang and Pardee, 2003; Kulis and Esteller, 2010). Moreover, coding sequence methylation can also control the gene activity by altering the nucleosome orientation inside the chromatin structure (Ehrlich, 2002; Luczak and Jagodziński, 2006). Additionally, the reduced methylation of different genes can propel the tumorigenesis of healthy cells inside the human body by escalating the activity of different oncogenes and thus differential methylation remains a promising target for epigenetic clinical decisions in cancer treatment (Issa, 2007). In this experiment, the promoter regions of all the PSMCs were found to be differentially methylated (less methylated) in LUAD tissues compared to the normal lung tissues (Figure 4). Thus, the overexpression of the PSMCs in LUAD tissues may be attributed to the less methylated regions in PSMC coding promoters. In a recent study, PSMC5 methylation has been linked to being negatively associated with colorectal cancer exacerbation (He et al., 2021). Furthermore, the evidence on the association between PSMC5 hypomethylation and LUAD patients’ poor OS and RFS reveals the potential of PSMCs in epigenetic-based therapeutic discovery for LUAD treatment (Supplementary Figure S3). Moreover, the differentially methylated circulating genes from samples like urine or blood of cancer patients can serve as a diagnostic marker for early-stage detection of lung cancer (Hong and Kim, 2021). In this experiment, several different regions of PSMC coding sequences have been found to have distinct methylation patterns across LUAD samples which along with the differential level of promoter methylation may aid in the noninvasive diagnosis of LUAD patients (Figure 4).
Somatic driver mutations are the major etiological factors in LUAD development. Hence, the optimum understanding of the genetic alterations in relevant genes and their relations to patients’ survival is paramount (Pao and Girard, 2011). Unsurprisingly, CNA contributes more to the oncogenic development and subsequent growth of healthy cells than other nonsynonymous mutations like point mutations (Zhao et al., 2004; Vikberg et al., 2017). In our study, all the PSMCs were predicted to have multiple somatic alteration events, including amplification, deep deletion, and splice which could promote the LUAD exacerbation by alternating the dosage of the translation products of the PSMCs inside the cells. In support of such assumptions, PSMC mutations in this study were associated with the poor OS of LUAD patients (Figure 5). What’s more, the presence of multiple missense mutations as evidenced in PSMC coding regions in LUAD patients may also influence the LUAD development and progression by producing non-functional, dysfunctional, or entirely no protein (Figure 5). Although the roles of PSMC gene mutations in cancer remain unstudied, multiple mutations in other proteasome family genes, i.e., PSMB5, PSMB6, and PSMB7, are associated with the myeloma cell survival (Shi et al., 2020). Apart from this, the prevalence of CNA events in different protein-coding genes can aid in the high-throughput diagnosis of lung cancer patients. For example, previously, different CNAs, including both loss and gain events in human chromosomes 3 and 6 have been successful in the high-throughput diagnosis of lung cancer within 44 months with an accuracy of 97% (Bowcock, 2014). Thus, this study’s observed alterations in PSMC genes could also be investigated in formulating PSMC-based diagnostic measures for the early stage and accurate screening of LUAD patients.
Furthermore, all the PSMC genes were found to be overexpressed at an earlier age in the LUAD patients. A significant increase in the expression levels of the genes was observed across different cancer stages and with advancing lymph node metastasis status (Figure 6). The expression of PSMC genes was negatively associated with OS and RFS of LUAD patients (Figure 7). These pieces of evidence suggest that PSMC-based diagnostic measures may serve as a practical diagnosis method that could allow the early-stage diagnosis and tracking of LUAD patients throughout the clinical courses.
Tumor-infiltrating immune cells play a crucial role in inhibiting cancer cell growth and different immune cells have been shown to improve the prognosis of lung cancer patients (Wang et al., 2019). Apart from this, the abundance of immune cells in cancer patients can also aid in tracking the patient’s status throughout the disease state and formulating immunotherapy for use during the clinical course (Bremnes et al., 2016). For example, previous studies have shown that the abundance of CD4+, CD8+ T cells, and neutrophils are prognostic factors in lung cancer (Woo et al., 2001; Eruslanov et al., 2014; Djenidi et al., 2015). In this study, a significant association between PSMCs expression and different immune cells infiltration including the ones mentioned earlier was observed in LUAD patients that might assist in propagating dual diagnosis along with the PSMC-based diagnosis method (Figure 7). On the other hand, the mutated form of the PSMC expression can alter the immune reactivity in the cancer microenvironment and worsen the prognosis of LUAD patients. Additionally, multiple PSMC mRNA expression levels were found to be positively and negatively correlated with the infiltration levels of different immunomodulators like CD274 (commonly known as programmed death-ligand 1; PD-L1) and indoleamine 2,3-dioxygenase 1 (IDO1) enzyme (Figure 8). PD-L1 is the most frequently found cell surface receptor in NSCLC, and its overexpression predicts poor survival of lung cancer patients. Additionally, PD-L1 checkpoint inhibition and anti-PD-L1 antibodies are the most widely studied immunotherapy approaches in lung cancer, as well as, anti-PD-L1 antibodies are approved by the Food and Drug Administration for IHC-based diagnosis of lung cancer (Steven et al., 2016; Ancevski Hunter et al., 2018). Moreover, IDO1 is a promising anticancer target for different cancer treatments including lung cancer whose function can be regulated by small candidate molecules and the process provides immune blockade opportunities outside the immune checkpoint inhibition and adoptive immune cell transfer (Du et al., 2019). Therefore, IDO1 and CD274 could guide the PSMC-based diagnostic methods and therapeutic option discovery for LUAD.
The co-expression analysis revealed that SLIRP is the top and highly co-expressed gene of PSMC1 which was shown to have prognostic roles in colorectal cancer (Salama et al., 2009). Among other selected top co-expressed genes, PSMA2 was found to promote colorectal cancer cell proliferation and NDUFS3 has been reported to be downregulated in the ovarian cancer cell and hypothesized to promote oncogenic development (Wang et al., 2013; Qi et al., 2021) (Figure 9). TIMM50, a gene found to be highly co-expressed with PSMC4, promotes tumorigenesis and acts as a prognostic indicator in NSCLC (Zhang et al., 2019). Moreover, in a study involving 129 colorectal cancer (CRC) patients, TIMM50 was discovered as a key regulator and prognostic marker of CRC (Sun et al., 2020). A pan-cancer analysis recently reported that CCDC137 plays a crucial role and acts as a prognostic marker in different forms of cancers (Guo et al., 2021). Given that the co-expressed genes are functionally related, all these shreds of evidence suggest that the PSMC family of genes might have an underlying mechanism in the oncogenic development of healthy lung cells.
Furthermore, all the positively co-expressed overlapping genes of PSMCs except PDCD45 in LUAD tissues were also found to be primarily associated with the LUAD development (Supplementary Figure S7). Gene ontology term analysis on these genes suggested that most are involved in DNA replication, controlling the cell cycle, and operating in the nucleus. The KEGG pathway analysis indicated that the genes are predominantly involved in different cancer pathways, development of bladder cancer, oncogenic virus infection pathways, and so on. These findings again signify that the PSMC family genes may be associated with LUAD development and progression since the deregulation of the activity of PSMCs’ co-expressed genes in LUAD tissues can result in the initiation of oncogenic processes (Byler et al., 2014). As a result, the co-expressed genes of the PSMCs could also be investigated in LUAD therapeutic and diagnostic measures discovery. However, further laboratory investigations are required on such assumptions. Finally, the overexpression pattern of PSMCs in LUAD tissues was evaluated in two independent small-scale (n = ∼20–60) microarray datasets (i.e., GSE116959 and GSE1037) while the mainstream analysis involved large-scale (n = ∼500–700) RNA sequencing data from LUAD samples of TCGA and GTEx cohorts. All the selected genes of this study also showed significant overexpression in the LUAD samples from the independent microarray datasets (Figure 11) (Supplementary Figure S8). The survival analysis of LUAD patients in relation to PSMC expression in the public dataset (GSE31210) also supported our initial analysis that overexpression of most of the PSMCs (PSMC2-5) is significantly and negatively associated with the OS of LUAD patients (Supplementary Figure S9).
Overall, this study demonstrated the differential expression of PSMCs in LUAD patients at both mRNA and protein levels. There was a significant association between PSMCs overexpression and LUAD patients’ clinical manifestation. Moreover, PSMCs overexpression was correlated to the poor OS and RFS of LUAD patients. All these pieces of evidence suggest that the transcriptomic and proteomic differential expression patterns of PSMCs could assist in the PSMC-based LUAD diagnosis. Moreover, their upregulation pattern in LUAD tissues may also be responsible for an elevated level of proteasome activities ultimately leading to the LUAD development and growth by aberrantly degrading the regulators of the cell cycle and apoptosis (Sterz et al., 2008; Park et al., 2018). Hence, the abnormal expression pattern of PSMCs in LUAD patients, irrespective of their demographic and clinical conditions, suggests that PSMCs could be a potential target for LUAD treatment option discovery. Moreover, all the genes of our interest showed variation in the methylation pattern of promoters and coding sequences between normal lung and LUAD tissues. Several missense and truncating mutations were reported in the PSMCs coding regions. Additionally, PSMC expression was found to be associated with different immune cells and immune modulators in LUAD microenvironment. Thus, the findings on genetic and epigenetic alterations and immune phenotypes of PSMCs may aid in preparing and increasing the precision of PSMC-based diagnostic and therapeutic approaches against LUAD. More specifically, out of the five selected PSMCs, PSMC4 showed the highest overexpression at the mRNA level (log2FC:0.8) and its overexpression was recorded in most of the LUAD cell lines. PSMC4 was also found to have the least methylated promoters in LUAD tissues among all the other PSMCs. Genetic alteration frequency was also the highest number for PSMC4 (2%) in LUAD tissues. Again, it also showed the highest HR (1.6) along with PSMC1 against the OS of LUAD patients. These indicate that PSMC4 might have the most prognostic power among the selected PSMCs in detecting the LUAD patients. However, such trajectories of our analysis require further investigation, and other PSMCs should also be investigated as they showed quite similar reports. Last, the functional enrichment analysis unveiled different co-expressed genes controlling biological processes during the cell cycle. Thus, the neighbor genes of PSMCs could also be investigated further while extending laboratory work on making PSMC-based diagnostic and therapeutic measures for LUAD patients. Overall, this study suggests that PSMCs and their transcriptional and translational products are efficient prognostic and therapeutic targets for LUAD diagnosis and treatment. The scientific findings of this study should aid in advancing further research on PSMC-based diagnostic and therapeutic development for LUAD and translating PSMCs into clinical practice.
Lastly, this study involved a large number of datasets to establish the prognostic and therapeutic potentials of PSMC1-5 in LUAD and most of the analysis was found to be significant. Later, the overexpression pattern and the association of these genes with LUAD patients’ survival rate were validated in a small-scale dataset and the inquiry was on par with our mainstream analysis. Moreover, this study provided multi-omics, i.e., genomic, transcriptomic, and proteomic overview of PSMC gene expression in LUAD prognosis. However, this study has some limitations, i.e., this study could not provide clearer insights into the molecular pathogenesis of PSMC genes in LUAD which requires further inspection. Alongside, though this study demonstrated the differential promoter methylation pattern of PSMC1-5 genes in LUAD, it could not testify whether the coding sequence of these genes is aberrantly methylated in LUAD tissues or not. Hereby, we warrant further laboratory research to extend the findings of this study which is currently underway by the authors.
Acknowledgments
Authors are thankful to Abu Rahad from the department of Biotechnology and Genetic Engineering, Jahangirnagar University, Dhaka, Bangladesh, for his supports in the data visualization of this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
Conceptualization and experimental design: MU. Experiment, analysis, and interpretation: MU, AM, and NI. Writing and review and editing: MU, AM, NI, SP, and BK. Supervision and funding acquisition: BK. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1I1A2066868), the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No: 2020R1A5A2019413), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HF20C0116), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HF20C0038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.935286/full#supplementary-material
References
- Allaire J. (2012). RStudio: Integrated Development Environment for R. Boston, MA, 165–171. 770(394). [Google Scholar]
- Ancevski Hunter K., Socinski M. A., Villaruz L. C. (2018). PD-L1 Testing in Guiding Patient Selection for PD-1/pd-L1 Inhibitor Therapy in Lung Cancer. Mol. Diagn Ther. 22 (1), 1–10. 10.1007/s40291-017-0308-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., et al. (2012). NCBI GEO: Archive for Functional Genomics Data Sets-Update. Nucleic acids Res. 41 (D1), D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashyam M. D. (2002). Understanding Cancer Metastasis. Cancer 94 (6), 1821–1829. 10.1002/cncr.10362 [DOI] [PubMed] [Google Scholar]
- Bowcock A. M. (2014). DNA Copy Number Changes as Diagnostic Tools for Lung Cancer. Thorax 69 (5), 496–497. 10.1136/thoraxjnl-2013-204681 [DOI] [PubMed] [Google Scholar]
- Bremnes R. M., Busund L.-T., Kilvær T. L., Andersen S., Richardsen E., Paulsen E. E., et al. (2016). The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-small Cell Lung Cancer. J. Thorac. Oncol. 11 (6), 789–800. 10.1016/j.jtho.2016.01.015 [DOI] [PubMed] [Google Scholar]
- Byler S., Goldgar S., Heerboth S., Leary M., Housman G., Moulton K., et al. (2014). Genetic and Epigenetic Aspects of Breast Cancer Progression and Therapy. Anticancer Res. 34 (3), 1071–1077. [PubMed] [Google Scholar]
- Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., Creighton C. J., Ponce-Rodriguez I., Chakravarthi B. V. S. K., et al. (2017). UALCAN: a Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19 (8), 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Fu L., Hu J., Guo G., Xie A. (2021). Silencing of PSMC2 Inhibits Development and Metastasis of Prostate Cancer through Regulating Proliferation, Apoptosis and Migration. Cancer Cell Int. 21, 235. 10.1186/s12935-021-01934-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Liu Y. X., Huang L. (2022). ImageGP: An Easy‐to‐use Data Visualization Web Server for Scientific Researchers. iMeta 1, e5. 10.1002/imt2.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. S., Seol W., Moore D. D. (1996). A Component of the 26S Proteasome Binds on Orphan Member of the Nuclear Hormone Receptor Superfamily. J. Steroid Biochem. Mol. Biol. 56 (1–6 Spec), 23–30. 10.1016/0960-0760(95)00220-0 [DOI] [PubMed] [Google Scholar]
- Devarakonda S., Govindan R. (2019). Untangling the Evolutionary Roots of Lung Cancer. Nat. Commun. 10 (1), 2979. 10.1038/s41467-019-10879-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Li Y., Fan H., Xu W., Gao R., Bai S., et al. (2019). Knockdown of PSMC3IP Suppresses the Proliferation and Xenografted Tumorigenesis of Hepatocellular Carcinoma Cell. J Cell. Biochem. 120, 5449–5458. 10.1002/jcb.27824 [DOI] [PubMed] [Google Scholar]
- Djenidi F., Adam J., Goubar A., Durgeau A., Meurice G., de Montpréville V., et al. (2015). CD8+CD103+ Tumor-Infiltrating Lymphocytes Are Tumor-specific Tissue-Resident Memory T Cells and a Prognostic Factor for Survival in Lung Cancer Patients. J. I. 194 (7), 3475–3486. 10.4049/jimmunol.1402711 [DOI] [PubMed] [Google Scholar]
- Du Q., Feng X., Wang Y., Xu X., Zhang Y., Qu X., et al. (2019). Discovery of Phosphonamidate Ido1 Inhibitors for the Treatment of Non-small Cell Lung Cancer. Eur. J. Med. Chem. 182, 111629. 10.1016/j.ejmech.2019.111629 [DOI] [PubMed] [Google Scholar]
- Ehrlich M. (2002). DNA Methylation in Cancer: Too Much, but Also Too Little. Oncogene 21 (35), 5400–5413. 10.1038/sj.onc.1205651 [DOI] [PubMed] [Google Scholar]
- Eruslanov E. B., Bhojnagarwala P. S., Quatromoni J. G., Stephen T. L., Ranganathan A., Deshpande C., et al. (2014). Tumor-associated Neutrophils Stimulate T Cell Responses in Early-Stage Human Lung Cancer. J. Clin. Invest. 124 (12), 5466–5480. 10.1172/jci77053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Watanabe T. K., Tanaka K., Slaughter C. A., DeMartino G. N. (1996). cDNA Cloning of P42, a Shared Subunit of Two Proteasome Regulatory Proteins, Reveals a Novel Member of the AAA Protein Family. FEBS Lett. 387 (2–3), 184–188. 10.1016/0014-5793(96)00489-9 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal 6 (269), pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., et al. (2004). Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 5 (10), R80–R86. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., et al. (2019). The UCSC Xena Platform for Public and Private Cancer Genomics Data Visualization and Interpretation. biorxiv 1, 326470. 10.1101/326470 [DOI] [Google Scholar]
- Gu Z. C., Enenkel C. (2014). Proteasome Assembly. Cell. Mol. Life Sci. 71 (24), 4729–4745. 10.1007/s00018-014-1699-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Li B., Lu Z., Liang H., Yang H., Chen Y., et al. (2021). CCDC137 Is a Prognostic Biomarker and Correlates with Immunosuppressive Tumor Microenvironment Based on Pan-Cancer Analysis. Front. Mol. Biosci. 8, 674863. 10.3389/fmolb.2021.674863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Yang X., Huang L., Zhou L., Zhang S., Sun J., et al. (2021). PSMC5 Promotes Proliferation and Metastasis of Colorectal Cancer by Activating Epithelial–Mesenchymal Transition Signaling and Modulating Immune Infiltrating Cells. Front. cell Dev. Biol. 9, 657917. 10.3389/fcell.2021.657917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H., Meirelles G. V., da Silva F. R., Telles G. P., Minghim R. (2015). InteractiVenn: a Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinforma. 16 (169), 1–7. 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch F. R., Scagliotti G. V., Mulshine J. L., Kwon R., Curran W. J., Wu Y.-L., et al. (2017). Lung Cancer: Current Therapies and New Targeted Treatments. Lancet 389 (10066), 299–311. 10.1016/s0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- Hong Y., Kim W. J. (2021). DNA Methylation Markers in Lung Cancer. Cg 22 (2), 79–87. 10.2174/1389202921999201013164110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle J., Tan K. H., Fisher E. M. C. (1997). Localization of Genes Encoding Two Human One-Domain Members of the AAA Family: PSMC5 (The Thyroid Hormone Receptor-Interacting Protein, TRIP1) and PSMC3 (The Tat-Binding Protein, TBP1). Hum. Genet. 99 (2), 285–288. 10.1007/s004390050356 [DOI] [PubMed] [Google Scholar]
- Issa J.-P. J. (2007). DNA Methylation as a Therapeutic Target in Cancer. Clin. Cancer Res. 13 (6), 1634–1637. 10.1158/1078-0432.ccr-06-2076 [DOI] [PubMed] [Google Scholar]
- Jones M. H., Virtanen C., Honjoh D., Miyoshi T., Satoh Y., Okumura S., et al. (2004). Two Prognostically Significant Subtypes of High-Grade Lung Neuroendocrine Tumours Independent of Small-Cell and Large-Cell Neuroendocrine Carcinomas Identified by Gene Expression Profiles. Lancet 363 (9411), 775–781. 10.1016/s0140-6736(04)15693-6 [DOI] [PubMed] [Google Scholar]
- Kao T.-J., Wu C.-C., Phan N. N., Liu Y.-H., Ta H. D. K., Anuraga G., et al. (2021). Prognoses and Genomic Analyses of Proteasome 26S Subunit, ATPase (PSMC) Family Genes in Clinical Breast Cancer. Aging 13 (14), 17970. 10.18632/aging.203345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A., Kosinski M., Biecek P., Fabian S. (2017). Package ‘survminer’. Drawing Survival Curves Using ‘ggplot2’(R Package Version 03 1). [Google Scholar]
- Kim C., Kim D. S., Nam D., Kim S.-H., Shim B. S., Ahn K. S. (2015). Melittin Exerts Antitumorigenic Effects in Human MM1.S Multiple Myeloma Cells through the Suppression of AKT/mTOR/S6K1/4E-BP1 Signaling Cascades. Orient Pharm. Exp. Med. 15 (1), 33–44. 10.1007/s13596-014-0172-4 [DOI] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., et al. (2016). Enrichr: a Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 44 (W1), W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M., Esteller M. (2010). DNA Methylation and Cancer. Adv. Genet. 70, 27–56. 10.1016/b978-0-12-380866-0.60002-2 [DOI] [PubMed] [Google Scholar]
- Li X., Sun Y., Huang S., Chen Y., Chen X., Li M., et al. (2019). Inhibition of AZIN2-Sv Induces Neovascularization and Improves Prognosis after Myocardial Infarction by Blocking Ubiquitin-dependent Talin1 Degradation and Activating the Akt Pathway. EBioMedicine 39, 69–82. 10.1016/j.ebiom.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. (2003). Analysing Differential Gene Expression in Cancer. Nat. Rev. Cancer 3 (11), 869–876. 10.1038/nrc1214 [DOI] [PubMed] [Google Scholar]
- Liu C.-J., Hu F.-F., Xia M.-X., Han L., Zhang Q., Guo A.-Y. (2018). GSCALite: a Web Server for Gene Set Cancer Analysis. Bioinformatics 34 (21), 3771–3772. 10.1093/bioinformatics/bty411 [DOI] [PubMed] [Google Scholar]
- Liu Y., Chen H., Li X., Zhang F., Kong L., Wang X., et al. (2021). PSMC2 Regulates Cell Cycle Progression through the P21/cyclin D1 Pathway and Predicts a Poor Prognosis in Human Hepatocellular Carcinoma. Front. Oncol. 11, 542. 10.3389/fonc.2021.607021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak M. W., Jagodziński P. P. (2006). The Role of DNA Methylation in Cancer Development. Folia Histochem Cytobiol. 44 (3), 143–154. [PubMed] [Google Scholar]
- Manasanch E. E., Orlowski R. Z. (2017). Proteasome Inhibitors in Cancer Therapy. Nat. Rev. Clin. Oncol. 14 (7), 417–433. 10.1038/nrclinonc.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Leon L., Gautier M., Allan R., Ilié M., Nottet N., Pons N., et al. (2019). The Nuclear Hypoxia-Regulated NLUCAT1 Long Non-coding RNA Contributes to an Aggressive Phenotype in Lung Adenocarcinoma through Regulation of Oxidative Stress. Oncogene 38 (46), 7146–7165. 10.1038/s41388-019-0935-y [DOI] [PubMed] [Google Scholar]
- Myers D. J., Wallen J. M. (2022). in StatPearls [Internet] (Treasure Island (FL): StatPearls Publishing; ). [Updated 2021 Sep 10].Lung Adenocarcinoma [Google Scholar]
- National Center for Biotechnology Information (NCBI) (2022). PSMC1 Proteasome (Prosome, Macropain) 26S Subunit, ATPase 1. Accessed from: https://www.ncbi.nlm.nih.gov/gene/5700 .(Accessed on April 20, 2022). [Google Scholar]
- Oberndorfer F., Müllauer L. (2018). Molecular Pathology of Lung Cancer: Current Status and Perspectives. Curr. Opin. Oncol. 30 (2), 69–76. 10.1097/cco.0000000000000429 [DOI] [PubMed] [Google Scholar]
- Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R., et al. (2012). Identification of Genes Upregulated in ALK-Positive and EGFR/KRAS/ALK-negative Lung Adenocarcinomas. Cancer Res. 72 (1), 100–111. 10.1158/0008-5472.can-11-1403 [DOI] [PubMed] [Google Scholar]
- Pao W., Girard N. (2011). New Driver Mutations in Non-small-cell Lung Cancer. lancet Oncol. 12 (2), 175–180. 10.1016/s1470-2045(10)70087-5 [DOI] [PubMed] [Google Scholar]
- Papatheodorou I., Fonseca N. A., Keays M., Tang Y. A., Barrera E., Bazant W., et al. (2018). Expression Atlas: Gene and Protein Expression across Multiple Studies and Organisms. Nucleic acids Res. 46 (D1), D246–D251. 10.1093/nar/gkx1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. E., Miller Z., Jun Y., Lee W., Kim K. B. (2018). Next-generation Proteasome Inhibitors for Cancer Therapy. Transl. Res. 198, 1–16. 10.1016/j.trsl.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontén F., Jirström K., Uhlen M. (2008). The Human Protein Atlas—A Tool for Pathology. J. Pathology A J. Pathological Soc. G. B. Irel. 216 (4), 387–393. 10.1002/path.2440 [DOI] [PubMed] [Google Scholar]
- Qi J., Hu Z., Liu S., Li F., Wang S., Wang W., et al. (2021). Comprehensively Analyzed Macrophage-Regulated Genes Indicate that PSMA2 Promotes Colorectal Cancer Progression. Front. Oncol. 10, 618902. 10.3389/fonc.2020.618902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Wang W., An F., Huang W., Ding J. (2019). PSMC2 Is Up-Regulated in Pancreatic Cancer and Promotes Cancer Cell Proliferation and Inhibits Apoptosis. J. Cancer 10 (20), 4939–4946. 10.7150/jca.27616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru B., Wong C. N., Tong Y., Zhong J. Y., Zhong S. S. W., Wu W. C., et al. (2019). TISIDB: an Integrated Repository Portal for Tumor-Immune System Interactions. Bioinformatics 35 (20), 4200–4202. 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- Salama P., Phillips M., Grieu F., Morris M., Zeps N., Joseph D., et al. (2009). Tumor-infiltrating FOXP3+ T Regulatory Cells Show Strong Prognostic Significance in Colorectal Cancer. Jco 27 (2), 186–192. 10.1200/jco.2008.18.7229 [DOI] [PubMed] [Google Scholar]
- Senosain M.-F., Massion P. P. (2020). Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front. Oncol. 10, 349. 10.3389/fonc.2020.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-X., Zhu Y. X., Bruins L. A., Bonolo de Campos C., Stewart W., Braggio E., et al. (2020). Proteasome Subunits Differentially Control Myeloma Cell Viability and Proteasome Inhibitor Sensitivity. Mol. Cancer Res. 18 (10), 1453–1464. 10.1158/1541-7786.mcr-19-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H., Irie K., Ninomiya-Tsuji J., Goebl M., Taniguchi T., Matsumoto K. (1992). New Human Gene Encoding a Positive Modulator of HIV Tat-Mediated Transactivation. Nature 357 (6380), 700–702. 10.1038/357700a0 [DOI] [PubMed] [Google Scholar]
- Song M., Wang Y., Zhang Z., Wang S. (2017). PSMC2 Is Up-Regulated in Osteosarcoma and Regulates Osteosarcoma Cell Proliferation, Apoptosis and Migration. Oncotarget 8, 933–953. 10.18632/oncotarget.13511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterz J., Metzler I. v., Hahne J.-C., Lamottke B., Rademacher J., Heider U., et al. (2008). The Potential of Proteasome Inhibitors in Cancer Therapy. Expert Opin. investigational drugs 17 (6), 879–895. 10.1517/13543784.17.6.879 [DOI] [PubMed] [Google Scholar]
- Steven A., Fisher S. A., Robinson B. W. (2016). Immunotherapy for Lung Cancer. Respirology 21 (5), 821–833. 10.1111/resp.12789 [DOI] [PubMed] [Google Scholar]
- Su C. C., Yeh S. H., Chen D. S., Chen P. J., Jou Y. S. (2007). OncoDB.HCC: an Integrated Oncogenomic Database of Hepatocellular Carcinoma Revealed Aberrant Cancer Target Genes and Loci. Nucleic Acids Res. 35 (Suppl. l_1), D727–D731. 10.1093/nar/gkl845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Wang J., Zhu Y. F., Li Z. Y., Xiang J. B., Chen Z. Y., et al. (2020). Prognostic Value of TIMM50 Expression in Colorectal Cancer. Archives Med. Sci. 16 (1), 1. 10.5114/aoms.2020.94487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 71, 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tanahashi N., Suzuki M., Fujiwara T., Takahashi E.-i., Shimbara N., Chung C. H., et al. (1998). Chromosomal Localization and Immunological Analysis of a Family of Human 26S Proteasomal ATPases. Biochem. Biophysical Res. Commun. 243 (1), 229–232. 10.1006/bbrc.1997.7892 [DOI] [PubMed] [Google Scholar]
- Tang Z., Kang B., Li C., Chen T., Zhang Z. (2019). GEPIA2: an Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic acids Res. 47 (W1), W556–W560. 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikberg A.-L., Vooder T., Lokk K., Annilo T., Golovleva I. (2017). Mutation Analysis and Copy Number Alterations of KIF23 in Non-small-cell Lung Cancer Exhibiting KIF23 Over-expression. Ott 10, 4969–4979. 10.2147/ott.s138420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Cheng X., Fu Z., Zhou C., Lu W., Xie X. (2013). Reduced Expression of NDUFS3 and its Clinical Significance in Serous Ovarian Cancer. Int. J. Gynecol. Cancer 23 (4), 622–629. 10.1097/IGC.0b013e318287a90d [DOI] [PubMed] [Google Scholar]
- Wang S.-s., Liu W., Ly D., Xu H., Qu L., Zhang L. (2019). Tumor-infiltrating B Cells: Their Role and Application in Anti-tumor Immunity in Lung Cancer. Cell Mol. Immunol. 16 (1), 6–18. 10.1038/s41423-018-0027-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xiong H., Zuo Y., Hu S., Zhu C., Min A. (2022). PSMC2 Knockdown Inhibits the Progression of Oral Squamous Cell Carcinoma by Promoting Apoptosis via PI3K/Akt Pathway. Cell Cycle 21 (5), 477–488. 10.1080/15384101.2021.2021722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., Chang W., Wickham M. H. (2016). Package ‘ggplot2’. Create Elegant Data Visualisations Using the Grammar of Graphics. Version 2 (1), 1–89. [Google Scholar]
- Woo E. Y., Chu C. S., Goletz T. J., Schlienger K., Yeh H., Coukos G., et al. (2001). Regulatory CD4(+)CD25(+) T Cells in Tumors from Patients with Early-Stage Non-small Cell Lung Cancer and Late-Stage Ovarian Cancer. Cancer Res. 61 (12), 4766–4772. [PubMed] [Google Scholar]
- Zhang J. Y., Shi K. Z., Liao X. Y., Li S. J., Bao D., Qian Y., et al. (2021). The Silence of PSMC6 Inhibits Cell Growth and Metastasis in Lung Adenocarcinoma. Biomed. Res. Int. 2021, 9922185. 10.1155/2021/9922185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Han S., Zhou H., Cai L., Li J., Liu N., et al. (2019). TIMM50 Promotes Tumor Progression via ERK Signaling and Predicts Poor Prognosis of Non‐small Cell Lung Cancer Patients. Mol. Carcinog. 58 (5), 767–776. 10.1002/mc.22969 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Cao X., Li P., Fan Y., Zhang L., Li W., et al. (2020). PSMC6 Promotes Osteoblast Apoptosis through Inhibiting PI3K/AKT Signaling Pathway Activation in Ovariectomy‐induced Osteoporosis Mouse Model. J. Cell Physiol. 235, 5511–5524. 10.1002/jcp.29261 [DOI] [PubMed] [Google Scholar]
- Zhao X., Li C., Paez J. G., Chin K., Jänne P. A., Chen T.-H., et al. (2004). An Integrated View of Copy Number and Allelic Alterations in the Cancer Genome Using Single Nucleotide Polymorphism Arrays. Cancer Res. 64 (9), 3060–3071. 10.1158/0008-5472.can-03-3308 [DOI] [PubMed] [Google Scholar]
- Zheng X., Li Y., Ma C., Zhang J., Zhang Y., Fu Z., et al. (2020). Independent Prognostic Potential of GNPNAT1 in Lung Adenocarcinoma. Biomed. Res. Int. 2020, 8851437. 10.1155/2020/8851437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wang Y., Wang D., Wan J. (2022). PSMC2 Is Overexpressed in Glioma and Promotes Proliferation and Anti-apoptosis of Glioma Cells. World J. Surg. Onc 20 (84), 1. 10.1186/s12957-022-02533-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.