Abstract
Background
Heart failure is currently divided into three main forms, HFrEF, HFpEF, and HFmrEF, but its etiology is diverse and highly heterogeneous. Many studies reported a variety of novel subgroups in heart failure patients, with unsupervised machine learning methods. The aim of this scoping review is to provide insights into how these techniques can diagnose and manage HF faster and better, thus providing direction for future research and facilitating its routine use in clinical practice.
Methods
The review was performed following PRISMA-SCR guideline. We searched the PubMed database for eligible publications. Studies were included if they defined new subgroups in HF patients using clustering analysis methods, and excluded if they are (1) Reviews, commentary, or editorials, (2) Studies not about defining new sub-types, or (3) Studies not using unsupervised algorithms. All study screening and data extraction were conducted independently by two investigators and narrative integration of data extracted from included studies was performed.
Results
Of the 498 studies identified, 47 were included in the analysis. Most studies (61.7%) were published in 2020 and later. The largest number of studies (46.8%) coming from the United States, and most of the studies were authored and included in the same country. The most commonly used machine learning method was hierarchical cluster analysis (46.8%), the most commonly used cluster variable type was comorbidity (61.7%), and the least used cluster variable type was genomics (12.8%). Most of the studies used data sets of less than 500 patients (48.9%), and the sample size had negative correlation with the number of clustering variables. The majority of studies (85.1%) assessed the association between cluster grouping and at least one outcomes, with death and hospitalization being the most commonly used outcome measures.
Conclusion
This scoping review provides an overview of recent studies proposing novel HF subgroups based on clustering analysis. Differences were found in study design, study population, clustering methods and variables, and outcomes of interests, and we provided insights into how these studies were conducted and identify the knowledge gaps to guide future research.
Keywords: heart failure, subtype, machine learning, clustering analysis, scoping review
Introduction
Heart failure (HF) is the serious manifestation and terminal stage of many cardiovascular diseases, with a high level of mortality and readmission rate (1). The global prevalence of HF is about 26 million, and with the aggravation of population aging and the increase of survival rate of acute coronary syndrome (ACS), the prevalence of HF is increasing continuously (2). However, the existing treatment measures are only symptomatic support treatments to improve symptoms, but cannot completely reverse the course of disease. One of the reasons for this phenomenon is that the current HF subpopulation cannot fully integrate the heterogeneity of HF clinical manifestations and progression, which further aggravates the serious consequences caused by inadequate or even inaccurate phenotypic classification.
In previous guidelines for heart failure, heart failure was classified according to the cut-off point of LVEF—heart failure with reduced ejection fraction (HFrEF): HF with LVEF ≤ 40%; Heart failure with preserved ejection fraction (HFpEF): HF with LVEF ≥ 50%; Heart failure with intermediate ejection fraction (HFmrEF): HF with LVEF > 40% and L VEF < 50% (3). The new guideline proposed a new and revised classification of HF according to LVEF: HF with improved ejection fraction (HFimpEF): symptomatic HF with a baseline LVEF ≤ 40%, a ≥ 10 points increase from baseline LVEF, and a second measurement of LVEF > 40%. When classifying heart failure based on LVEF, previous guidelines have used HFrEF and HFpEF, but for the types of heart failure with EF values between 40 and 49%, there are different terms used, and there is no uniform standard. In the new classification, patients with normalized EF may have decreased EF after drug treatment was discontinued, meaning that although EF improved, cardiac structure and function did not (4). Although large number of studies have analyzed and summarized the structural and functional characteristics of cardiac cells, intercellular excitation conduction pathway, and cellular inflammation degree of patients in each subtype under this classic classification, there is still a situation of lack of effective treatment and limited personalized medical care, which urgently requires more accurate and detailed grouping strategies (5, 6). The complexity of the development of heart failure is difficult to explain with the emphasis on symptoms and signs in the previous diagnostic classification. We believe that the new subtype will give new directions in the interpretation of heterogeneity and treatment selection. The introduction of subgroups of patients with homogeneous characteristics is helpful to treat patients according to their clinical and pathophysiological characteristics, reduces the complexity of the cross influence of data characteristics of different dimensions during the treatment of patients, and plays a role in improving the treatment and prognosis (7).
Machine learning (ML) has achieved good accuracy in early diagnosis, clinical classification and risk factor prediction of patients with HF (8, 9). However, because of the black box feature of the algorithm, we cannot learn from the classification process of the algorithm. Unsupervised machine learning, specifically clustering analysis, is used to find the similar or different features between patients groups, and identify subgroups with homogeneous features. Clustering studies have certain advantages in characterizing, classifying or treating patients differently. Clustering algorithms commonly are performed in a static way with baseline data and/or outcome data. They are useful to answer descriptive questions (10, 11). In the early attempts, unsupervised clustering analysis algorithms were used on clinical laboratory indexes and demographic data characteristics of patients with heart failure to make homogeneous inductive groups (12, 13). In recent studies, researchers also used echocardiography, genomics and comorbidity characteristics to explore more grouping strategies (14). Without knowing the outcomes information (i.e., unsupervised learning), clustering analysis can comprehensively reflect the association between new subgroups and heart failure outcomes and other prognostic indicators.
There are wide variations in studies defining new heart failure subgroups, in study design, statistical methods, and reporting of outcomes, which makes comparing and summarizing results from different studies very difficult. Therefore, it is necessary to conduct a scope review to summarize the current practice in studies on the new subgroups of heart failure, clarify the limitations and provide direction and planning for the future research. At present, some researchers have discussed the application of machine learning in heart failure subtypes. Banerjee et al. included 15 studies published up to 2015, and compared the symptoms of cardiovascular diseases such as ACS, myocardial infarction (MI) and heart failure (HF) (15); In addition, Banerjee and others evaluated the subtype definition and risk prediction of ML in HF, ACS and AF (Atrial Fibrillation), and systematically reviewed them (15). However, in their research, the definitions of heart failure and subgroups are only a part of the research, and the clustering variables concerned are not comprehensive, and the included research is up to December, 2019 at the latest. Therefore, it is necessary to define the scope with a specific focus on the subtype classification in heart failure, and fully incorporating the latest research reports. This scoping review will integrate the current evidence on subtype classification of heart failure reported in the existing literature to provide a reference for clinicians and community health care workers to manage HF better, and identify the knowledge gaps to point out the direction for future research.
Methods
This scoping review followed the Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) guideline (16), and a competed PRISMA-ScR checklist was provided in Supplementary material. A study protocol was designed by a senior author and agreed by all authors, and this protocol was not registered or published.
Literature search
We performed a search in PubMed to identify primary studies on discovery of new HF sub-types by using clustering analysis. The search strategy contained 3 modules: the HF module, the algorithm module, and the sub-type module, and a filter of publication time till 31st December 2021 (see Supplementary material Search Strategy).
Eligibility criteria
Studies were included if they defined new subgroups in HF patients using clustering analysis methods. Exclusion criteria were: (1) Reviews, commentary, or editorials, (2) Studies not about defining new sub-types, (3) Studies not using unsupervised algorithms.
Study selection
Titles and abstracts were independently scanned by one of the two authors and checked by the other, to identify potentially eligible articles, which were then assessed with full texts for final inclusion. Disagreements were resolved through discussion by the two reviewers, and a third author made the final decision when an agreement was not reached.
Data extraction
Data was collected on basic study characteristics including title, name of the first author, year of publication, country, and information and the analysis and results including study population, sample size, clustering method(s), types of clustering variables, and outcome(s). The data extraction form and data extracted in this study can be found in the Supplementary material. All included articles were reviewed and extracted by one of the two authors and double checked by the other. Disagreements were resolved through discussion, if necessary the final judgment was from a third reviewer.
Data synthesis
Data synthesis was performed with descriptive statistics and data visualization. Categorical variables were presented as counts and proportions, and continuous variables were presented with median and IQR. All the statistical analyses were performed with R version 3.6.1 and RStudio version 1.2.5001, and packages ggplot2, networkD3 (sankey diagram), ggparliament (parliament diagram), UpSetR (upset plot) and scatterpie (bubble chart).
Results
Search finding
A total of 498 studies were identified by the search strategy, all of them were screened for titles and abstracts, of which 446 were excluded at this stage. Fifty-two studies entered the stage of full-text reading to assess their qualification, and five of them were excluded, for reasons shown in Figure 1. In the end, 47 studies were included in the review.
FIGURE 1.
PRISMA flow diagram for study inclusion.
Characteristics of the included studies
Table 1 shows the basic characteristics of the included studies in this review. Among the 47 included studies, 23(48.9%) studies focused on patients with generalized HF (7, 13, 17–31), while the rest 24 studies focused on patients with specific categories of HF, among which 19 studies (40.4%) focused on patients with HFpEF (32–50), and the other 5 studies (14, 51–54) (10.6%) focused on HFrEF. Of these studies, only one (2.1%) was published before 2010 (55) and four (8.5%) were published between 2010 and 2014 (19, 20, 54, 56). Most of the research was published after 2015, and the research after 2020 accounted for 61.7% (32, 34, 35, 37, 38, 40, 42–44), as shown in Figure 2.
TABLE 1.
Descriptive statistics of study characteristics.
| Characteristics | Studies, n (%)/M(Q1–Q3) |
| HF subtype | |
| HF | 23 (48.9%) |
| HFpEF | 19 (40.4%) |
| HFrEF | 5 (10.6%) |
| Years published | |
| <2010 | 1 (2.1%) |
| 2010–2014 | 4 (8.5%) |
| 2015–2019 | 13 (27.7%) |
| ≥2020 | 29 (61.7%) |
| Author country | |
| Asia | 5 (10.6%) |
| Europe | 18 (38.3%) |
| America | 23 (48.9%) |
| Oceania | 1 (2.1%) |
| Data source | |
| Asia | 4 (8.5%) |
| Europe | 14 (29.8%) |
| America | 21 (44.7%) |
| Oceania | 1 (2.1%) |
| Multinational | 7 (14.9%) |
| Data types | |
| EHR | 24 (51.1%) |
| RCT | 9 (19.1%) |
| Disease registries | 8 (17.0%) |
| Observational data | 3 (6.4%) |
| Claims data | 1 (2.1%) |
| EHR and claims data | 1 (2.1%) |
| EHR, RCT, and registries data | 1 (2.1%) |
| Method | |
| K-Means/Medoids | 9 (19.1%) |
| LCA | 11 (23.4%) |
| Hierarchical | 22 (46.8%) |
| Hierarchical & K-Means/Medoids | 1 (2.1%) |
| SOM | 1 (2.1%) |
| Spectral | 1 (2.1%) |
| Mixture model-based | 2 (4.3%) |
| Sample size | 480 (301–1619) |
| Number of variables | 18 (11–47) |
| Number of clusters | 3 (3–6) |
| Variable type | |
| Demographic | 24 (51.1%) |
| Clinical | 25 (53.2%) |
| Laboratory | 21 (44.7%) |
| Imaging | 24 (51.1%) |
| Genetic | 6 (12.8%) |
| Symptoms and complaints | 18 (38.3%) |
| Comorbidities | 29 (61.7%) |
| Outcome | |
| Cross sectional | 7 (14.9%) |
| Mortality | 36 (76.6%) |
| Hospitalisation | 27 (57.4%) |
| Other events | 14 (29.8%) |
| External validation | |
| Yes | 6 (12.8%) |
| No | 41 (87.2%) |
FIGURE 2.
Number of publication per year by HF sub-types.
In all the included studies, the corresponding authors were from 13 different countries, including the United States (22, 46.8%) (18–22, 24, 26–28, 33, 36, 37, 40, 41, 44, 48, 53, 54, 56–59), the Netherlands (5, 10.6%) (23, 45, 47, 55, 60), France (5, 10.6%) (13, 17, 32, 43, 51), Spain (3, 6.4%) (25, 34, 38), China (3, 6.4%) (7, 35, 49) and Japan (2, 4.3%) (31). Australia (52), Germany (30), Italy (14), Poland (39), Switzerland (46), Canada (50) and the United Kingdom (29) each had only one study (1, 2.1%). The research data were from a single country in 40 studies (85.1%), and the rest 7 studies (14.9%) were performed with multinational data (13, 17, 24, 28, 37, 43, 60). The relationship between data sources and corresponding authors is shown in Figure 3. We further classify them according to their continents, and find that the highest number of authors and participants are from America and Europe, followed by Asia and Oceania, as shown in Table 1.
FIGURE 3.
Relationship between data sources and corresponding authors.
We analyzed the types of data source, the results showed that in all included in the article, there are 24 articles (51.1%) using EHR data, 9 articles (19.1%) using RCT research data, 8 articles (17.0%) using disease registration data, 3 article (6.4%) using the observational data, 1 article (2.1%) using the claims data. In addition, one study used EHR data and Claims data simultaneously, and another study used EHR data, RCT data and registries data simultaneously.
In addition, of the 47 included studies, only 6 (12.8%) were externally validated, while the remaining 41 studies (87.2%) were not.
Types of clustering methods in the included studies
The clustering methods were categorized into six main types, and the usage of each type of methods in the included studies are shown in Figure 4 and Table 1. The most commonly used ML method was hierarchical clustering method (32–33, 35, 38–41, 43, 44), accounting for 46.8% (22/47) of all studies, followed by latent class analysis (11, 23.4%) (7, 17, 24, 26, 29, 30, 32, 36, 37, 47, 55) and K-Means/Medoids (9, 19.1%) (13, 28, 31, 34, 42, 50, 52, 53, 60), and two studies used mixture model-based approach (4.3%) (46, 58). The least commonly used methods were spectral (49), self-organizing map (23) and composite of hierarchical and K-Means/Medoids (27).
FIGURE 4.
Types of machine learning methods used in identifying HF subgroups.
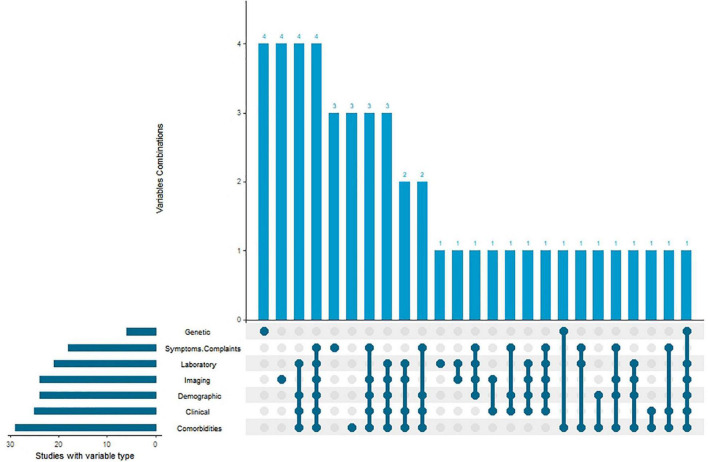
Types of clustering variables used in the included studies
We divide all the variables used by the institute for unsupervised cluster analysis into seven categories: demographic data (such as gender, age, education level, etc.), clinical data (such as heart rate, respiratory rate, etc.), laboratory data, image features (such as LEVF, etc.), genetic data, clinical symptoms and complications, and comorbidities. We have sorted out the frequency with which these variable types are used and whether they are jointly used in cluster analysis, as shown in Table 1 and Figure 5. As far as the frequency of variable types is concerned, comorbidities are the most frequently used in these studies 29(61.7%), and the clinical data, imaging data, demographic data and laboratory data are almost the same, which are 25(53.2%), 24(51.1%), 24(51.1%) and 21(44.7%) respectively. Among them, gene data, symptoms and complications data, image data, laboratory data and complications data are used alone in the process of sub-grouping in some studies. Most studies combine multiple data types to make a new subgroup classification of heart failure.
FIGURE 5.
Types of clustering variables used in identifying HF subgroups.
Sample size, number of clustering variables, and number of clusters
The sample sizes in the derivation of subgroups ranged from 63 to 318,384. In all 47 studies, the median sample size was 480, of which 23 studies (48.9%) included data sets of more than 500 people, and only 2 studies (4.3%) had a sample size of less than 100 people. The number of clustering variables involved in the research also varies widely, the research with the least variables was using only 7 clinical, laboratory or imaging indicators, and the research with the most variables was using 13,000 genes for clustering analysis to determine their molecular subgroups. The number of clusters obtained in most studies ranged from 2 to 7 (97.9%), with a median of 3, of which 16 studies (34.0%) finally got 3 clusters, and only one study got 11 clusters. Figure 6 shows the relationship between the number of variables (X-axis, in log scale) and the number of clusters (Y-axis) identified in each study, and the sample size (in log scale) is presented as the radius of the bubble. The Spearman correlation was −0.14 between sample size and number of clustering variables, 0.15 between sample size and number of clusters, and 0.13 between number of clustering variables and number of clusters.
FIGURE 6.
Features of the clusters identified in the included studies.
Prognostic implications of the clusters proposed in the included studies
Many different outcomes were used to evaluate the prognostic implication of the identified new subgroups, thus they were classified into four categories: death, hospitalization, other events, and cross-sectional study (i.e., no prognostication was assessed). The most commonly used endpoints were death (36, 76.6%) and hospital (27, 57.4%) respectively, while in 7 studies (14.9%) no analysis was performed for prognostic implications. The outcomes evaluated in each study are shown in Figure 6.
Discussion
In this scoping review, we summarized the current research on identifying subgroups in HF patients with unsupervised machine learning methods. This type of studies increased quickly over past years, and there were 19 new publications in 2021. Differences were found in study design, study population, clustering methods and variables, and outcomes of interests, and we aimed to provide insights into how these studies were conducted and identify the knowledge gaps to guide future research.
Most of the studies were conducted by researchers from developed countries, or in geographic, from Europe and North America, which is not a surprising finding given their leading position in the field of biomedical and clinical researches. However, subgroups identified from these populations may have poor generalizability in other part of the world. Africa, South America, South and West Asia were under represented, since the data availability is limited in those areas. We also noticed that most researchers worked on data from their own country, and only a few studies were from multinational collaboration. Future research should consider combining datasets containing patients from different countries and regions, to investigate the potential application of the new subgroups worldwide.
With regarding to the clustering methods, in this scoping review, the most commonly used unsupervised machine learning algorithm is hierarchical clustering, followed by K-Means or K-Medoids. Hierarchical clustering has the advantage of not requesting predefined number of clusters, which is useful in exploring novel subgroups. Researchers have had long history in using K-Means or K-Medoids clustering in analyzing data, and these methods were considered as multivariate analysis before the term of machine learning getting its popularity (61, 62).
The novel HF subgroups can be defined with different types of variable, however, the application of subgroups also depends on how difficult these clustering variables can be collected. Obtaining demographic variables and underlying comorbidities is straightforward by asking the patient’s medical history at admission, which made them as the most frequently used variables in clustering analysis, and laboratory data and imaging data can also be obtained in routine examination after admission. However, genomics or proteomics data may need extra special examination methods, which are less common compared with other data types, so that genomics or proteomics are rarely used in the included studies, but this also heralds the great potential of genomics in revealing the prognosis of heart failure patients.
The implementation of machine learning methods relay on data in a large degree. In the included studies, 23 studies used datasets of less than 500 patients and only 15 studies used data sets of more than 1,000 patients. At the same time, the number of clustering variables is relatively big, sometime even higher than the sample size, which may lead to overfitting issues. A negative correlation was observed between the number of clustering variables and the sample size, which is not as expected. When more clustering variables are included in the analyses, researchers need to make sure the sample size is sufficient to get reliable results.
Some included studies did not evaluate the prognostic implication of the proposed subgroups, and we marked these studies as cross-sectional studies. Unsupervised cluster analysis does have obvious advantages in finding out the heterogeneity among patients, and the new subgroup is also more accurate in describing the symptoms and complications of patients, but not connecting with the prognosis means that it is limited in clinical application, so we hope that more researches will make a clear plan for the clinical endpoint of patients. In addition, we found that few studies have set the quality of life or daily behavior ability as the research endpoint, which may be due to the fact that similar endpoints need more detailed evaluation scales or multi-dimensional evaluation indicators, which are quite different in the nature of easy access compared with the outcome endpoints such as death or readmission.
Unlike traditional prediction models, which pay more attention to the prediction accuracy and absolute probability of having a specific event, clustering analysis focused on classifying complex and heterogeneous diseases and identifying people with similar clinical characteristics. Thus, subgroups identified with clustering analysis may have better explanation and clinical meaning than prediction models. With these novel subgroups, patients can be more accurately risk stratified by more simple and easily available clinical indicators, and then targeted treatment schemes can be formulated.
With the development of coronary intervention technology, more patients with coronary heart disease survive and develop into heart failure. Coupled with the aggravation of population aging, the number of patients with heart failure is increasing year by year (63, 64), there is a higher proportion of elderly patients among them, and the existence mode of comorbidity is more complicated, which is followed by the increase of medical expenses, mortality and hospitalization rate (65). More and more researchers are aware of the importance of comorbidity management. The study on comorbidity of patients with heart failure found that the number of participants suffering from diabetes, chronic kidney disease and atrial fibrillation was higher (66), and other common comorbidities included hypertension and chronic obstructive pulmonary diseases. Some studies showed that some comorbidities would change the disease phenotype of patients with heart failure, and even become the main cause of heart failure (67), whereas some studies reported that the comorbidity of patients with heart failure was more serious, which might be caused by heart failure. Diabetes, hypertension, etc., are also associated with worse clinical outcomes in other diseases. Therefore, it is of great significance to carry out more personalized management for patients with heart failure under different comorbidity modes. These common comorbidities are important variables in our included studies, and the emergence of new subgroups and new treatment standards have also brought about the improvement of clinical prognosis in these studies. In addition to the elderly patients with heart failure, recent studies have found that the prevalence of cardiovascular comorbidities in middle-aged patients with heart failure is also very high, compared with the elderly patients with heart failure (>85 years old) (68). In the included studies, the new grouping of patients based on comorbidity or combined with other types of data also provided reference for clinical treatment.
There are also some limitations of the current scoping review. First, when searching for eligible publications, we only performed the literature search in PubMed database. Some other databases such as scienceDirect, Embase, IEEE, Scopus, etc., were not searched specifically, since most of the relevant publications are covered by PubMed, and looking for more databases will only increase the duplicates and add unnecessary workload. Given this is a scope review rather than a systematic review, we strictly enforce this search strategy, and we are confident the results presented in this scoping review are not biased. Second, we only included publications in English, and excluded those in other languages, which may reduce the diversity of this scope review. Third, we did not evaluate the evaluation criteria and external validation of the novel subgroups, since they are seldom done in the included studies. We believe validation or replication of the proposed subgroups are essential before these subgroups will be used in clinical practice, and future studies should pay more attention to these analyses. At last, this scoping review is only a comprehensive description of the existing researches on subgroup identification in HF patients, thus no formal assessment on methodological quality (or risk of bias) or meta-analysis was performed in this review. These analyses are usually within a systematic review, and are beyond the scope of this scoping review. In future research, we plan to perform a systematic review on studies with similar subgroup definition and a meta-analysis on their prognostic performance.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
JS and HG screened the publications, extracted data, and prepared the first draft. WW conducted a rigorous review of the first and final drafts. XW and JD participated in the revision of the research protocol and data extraction form. KH and XG designed and conceptualized the research project, rigorously revised and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This work was supported by the Ministry of Industry and Information Technology of China (2020-01 03-3-1).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.895836/full#supplementary-material
References
- 1.Tomasoni D, Adamo M, Lombardi CM, Metra M. Highlights in heart failure. ESC Heart Fail. (2019) 6:1105–27. 10.1002/ehf2.12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savarese G, Lund LH. Global public health burden of heart failure. Cardiac Fail Rev. (2017) 3:7–11. 10.15420/cfr.2016:25:2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozkurt B, Coats AJS, Tsutsui H, Abdelhamid CM, Adamopoulos S, Albert N, et al. Universal definition and classification of heart failure: a report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure: endorsed by the Canadian Heart Failure Society, Heart Failure Association of India, Cardiac Society of Australia and New Zealand, and Chinese Heart Failure Association. Eur J Heart Fail. (2021) 23:352–80. 10.1002/ejhf.2115 [DOI] [PubMed] [Google Scholar]
- 4.Writing Committee Members, ACC/AHA Joint Committee Members. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure. J Card Fail. (2022) 28:e1–167. 10.1016/j.cardfail.2022.02.010 [DOI] [PubMed] [Google Scholar]
- 5.Warbrick I, Rabkin SW. Effect of the peptides relaxin, neuregulin, ghrelin and glucagon-like peptide-1, on cardiomyocyte factors involved in the molecular mechanisms leading to diastolic dysfunction and/or heart failure with preserved ejection fraction. Peptides. (2019) 111:33–41. 10.1016/j.peptides.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 6.Mishra S, Kass DA. Cellular and molecular pathobiology of heart failure with preserved ejection fraction. Nat Rev Cardiol. (2021) 18:400–23. 10.1038/s41569-020-00480-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng C, Han L, Tian J, Li J, He H, Han G, et al. Hierarchical management of chronic heart failure: a perspective based on the latent structure of comorbidities. ESC Heart Fail. (2022) 9:595–605. 10.1002/ehf2.13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Martinez S, Duchateau N, Erdei T, Fraser AG, Bijnens BH, Piella G. Characterization of myocardial motion patterns by unsupervised multiple kernel learning. Med Image Anal. (2017) 35:70–82. 10.1016/j.media.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 9.Motwani M, Dey D, Berman DS, Germano G, Achenbach S, Al-Mallah MH, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. (2017) 38:500–7. 10.1093/eurheartj/ehw188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Li Z, Xiong H, Gao X, Wu J, Wu S. Understanding and enhancement of internal clustering validation measures. IEEE Trans Cybern. (2013) 43:982–94. 10.1109/tsmcb.2012.2220543 [DOI] [PubMed] [Google Scholar]
- 11.Gravesteijn BY, Steyerberg EW, Lingsma HF. Modern learning from big data in critical care: primum non nocere. Neurocrit Care. (2022). [Epub ahead of print]. 10.1007/s12028-022-01510-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omar AMS, Narula S, Abdel Rahman MA, Pedrizzetti G, Raslan H, Rifaie O, et al. Precision phenotyping in heart failure and pattern clustering of ultrasound data for the assessment of diastolic dysfunction. JACC Cardiovasc Imaging. (2017) 10:1291–303. 10.1016/j.jcmg.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 13.Stienen S, Ferreira JP, Kobayashi M, Preud’homme G, Dobre D, Machu JL, et al. Enhanced clinical phenotyping by mechanistic bioprofiling in heart failure with preserved ejection fraction: insights from the MEDIA-DHF study (The Metabolic Road to Diastolic Heart Failure). Biomarkers. (2020) 25:201–11. 10.1080/1354750x.2020.1727015 [DOI] [PubMed] [Google Scholar]
- 14.Carluccio E, Pugliese NR, Biagioli P, Zuchi C, Lauciello R, Mengoni A, et al. Global longitudinal strain in heart failure with reduced ejection fraction: prognostic relevance across disease severity as assessed by automated cluster analysis. Int J Cardiol. (2021) 332:91–8. 10.1016/j.ijcard.2021.02.072 [DOI] [PubMed] [Google Scholar]
- 15.Banerjee A, Chen S, Fatemifar G, Zeina M, Lumbers RT, Mielke J, et al. Machine learning for subtype definition and risk prediction in heart failure, acute coronary syndromes and atrial fibrillation: systematic review of validity and clinical utility. BMC Med. (2021) 19:85. 10.1186/s12916-021-01940-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. (2018) 169:467–73. 10.7326/m18-0850 [DOI] [PubMed] [Google Scholar]
- 17.Tamisier R, Damy T, Bailly S, Davy JM, Verbraecken J, Lavergne F, et al. Adaptive servo ventilation for sleep apnoea in heart failure: the FACE study 3-month data. Thorax. (2022) 77:178–85. 10.1136/thoraxjnl-2021-217205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sethares KA, Chin E. Age and gender differences in physical heart failure symptom clusters. Heart Lung. (2021) 50:832–7. 10.1016/j.hrtlng.2021.07.001 [DOI] [PubMed] [Google Scholar]
- 19.Vanburen P, Ma J, Chao S, Mueller E, Schneider DJ, Liew CC. Blood gene expression signatures associate with heart failure outcomes. Physiol Genomics. (2011) 43:392–7. 10.1152/physiolgenomics.00175.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad T, Pencina MJ, Schulte PJ, O’Brien E, Whellan DJ, Piña IL, et al. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J Am Coll Cardiol. (2014) 64:1765–74. 10.1016/j.jacc.2014.07.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad T, Desai N, Wilson F, Schulte P, Dunning A, Jacoby D, et al. Clinical implications of cluster analysis-based classification of acute decompensated heart failure and correlation with bedside hemodynamic profiles. PLoS One. (2016) 11:e0145881. 10.1371/journal.pone.0145881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevaert AB, Tibebu S, Mamas MA, Ravindra NG, Lee SF, Ahmad T, et al. Clinical phenogroups are more effective than left ventricular ejection fraction categories in stratifying heart failure outcomes. ESC Heart Fail. (2021) 8:2741–54. 10.1002/ehf2.13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uszko-Lencer N, Janssen DJA, Gaffron S, Vanfleteren L, Janssen E, Werter C, et al. Clustering based on comorbidities in patients with chronic heart failure: an illustration of clinical diversity. ESC Heart Fail. (2022) 9:614–26. 10.1002/ehf2.13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Johantgen ME. A cross-cultural comparison of symptom reporting and symptom clusters in heart failure. J. Transcult Nurs. (2017) 28:372–80. 10.1177/1043659616651673 [DOI] [PubMed] [Google Scholar]
- 25.Yun S, Enjuanes C, Calero-Molina E, Hidalgo E, José N, Calvo E, et al. Effectiveness of telemedicine in patients with heart failure according to frailty phenotypes: insights from the iCOR randomised controlled trial. Eur J Intern Med. (2022) 96:49–59. 10.1016/j.ejim.2021.09.021 [DOI] [PubMed] [Google Scholar]
- 26.Park J, Moser DK, Griffith K, Harring JR, Johantgen M. Exploring symptom clusters in people with heart failure. Clin Nurs Res. (2019) 28:165–81. 10.1177/1054773817729606 [DOI] [PubMed] [Google Scholar]
- 27.Bose E, Radhakrishnan K. Using unsupervised machine learning to identify subgroups among home health patients with heart failure using telehealth. Comput Inform Nurs. (2018) 36:242–8. 10.1097/cin.0000000000000423 [DOI] [PubMed] [Google Scholar]
- 28.Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, et al. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur J Heart Fail. (2019) 21:74–85. 10.1002/ejhf.1333 [DOI] [PubMed] [Google Scholar]
- 29.Gulea C, Zakeri R, Quint JK. Model-based comorbidity clusters in patients with heart failure: association with clinical outcomes and healthcare utilization. BMC Med. (2021) 19:9. 10.1186/s12916-020-01881-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henneges C, Morbach C, Sahiti F, Scholz N, Frantz S, Ertl G, et al. Sex-specific bimodal clustering of left ventricular ejection fraction in patients with acute heart failure. ESC Heart Fail. (2022) 9:786–90. 10.1002/ehf2.13618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiuchi Y, Tanimoto S, Latif A, Urayama KY, Aoki J, Yahagi K, et al. Identifying novel phenotypes of acute heart failure using cluster analysis of clinical variables. Int J Cardiol. (2018) 262:57–63. 10.1016/j.ijcard.2018.03.098 [DOI] [PubMed] [Google Scholar]
- 32.Fayol A, Wack M, Livrozet M, Carves JB, Domengé O, Vermersch E, et al. Aetiological classification and prognosis in patients with heart failure with preserved ejection fraction. ESC Heart Fail. (2022) 9:519–30. 10.1002/ehf2.13717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi Y, Tremblay-Gravel M, Boralkar KA, Li X, Nishi T, Amsallem M, et al. Approaching higher dimension imaging data using cluster-based hierarchical modeling in patients with heart failure preserved ejection fraction. Sci Rep. (2019) 9:10431. 10.1038/s41598-019-46873-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arévalo-Lorido JC, Carretero-Gómez J, Aramburu-Bodas O, Grau-Amoros J, Torres-Cortada G, Camafort-Babkowski M. Blood pressure, congestion and heart failure with preserved ejection fraction among patients with and without type 2 diabetes mellitus. A cluster analysis approach from the observational registry DICUMAP. High Blood Press Cardiovasc Prev. (2020) 27:399–408. 10.1007/s40292-020-00405-x [DOI] [PubMed] [Google Scholar]
- 35.Gu J, Pan JA, Lin H, Zhang JF, Wang CQ. Characteristics, prognosis and treatment response in distinct phenogroups of heart failure with preserved ejection fraction. Int J Cardiol. (2021) 323:148–54. 10.1016/j.ijcard.2020.08.065 [DOI] [PubMed] [Google Scholar]
- 36.Kao DP, Lewsey JD, Anand IS, Massie BM, Zile MR, Carson PE, et al. Characterization of subgroups of heart failure patients with preserved ejection fraction with possible implications for prognosis and treatment response. Eur J Heart Fail. (2015) 17:925–35. 10.1002/ejhf.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen JB, Schrauben SJ, Zhao L, Basso MD, Cvijic ME, Li Z, et al. Clinical phenogroups in heart failure with preserved ejection fraction: detailed phenotypes, prognosis, and response to spironolactone. JACC Heart Fail. (2020) 8:172–84. 10.1016/j.jchf.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arévalo-Lorido JC, Carretero-Gómez J, Gómez-Huelgas R, Llácer P, Manzano L, Quesada Simón MA, et al. Comorbidities and their implications in patients with and without type 2 diabetes mellitus and heart failure with preserved ejection fraction. Findings from the rica registry. Int J Clin Pract. (2021) 75:e13661. 10.1111/ijcp.13661 [DOI] [PubMed] [Google Scholar]
- 39.Przewlocka-Kosmala M, Marwick TH, Dabrowski A, Kosmala W. Contribution of cardiovascular reserve to prognostic categories of heart failure with preserved ejection fraction: a classification based on machine learning. J Am Soc Echocardiogr. (2019) 32:604–15.e6. 10.1016/j.echo.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 40.Pandey A, Kagiyama N, Yanamala N, Segar MW, Cho JS, Tokodi M, et al. Deep-learning models for the echocardiographic assessment of diastolic dysfunction. JACC Cardiovasc Imaging. (2021) 14:1887–900. 10.1016/j.jcmg.2021.04.010 [DOI] [PubMed] [Google Scholar]
- 41.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. (2015) 131:269–79. 10.1161/circulationaha.114.010637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harada D, Asanoi H, Noto T, Takagawa J. Different pathophysiology and outcomes of heart failure with preserved ejection fraction stratified by K-means clustering. Front Cardiovasc Med. (2020) 7:607760. 10.3389/fcvm.2020.607760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrub F, Oger E, Bidaut A, Hage C, Charton M, Daubert JC, et al. Heart failure with preserved ejection fraction: a clustering approach to a heterogenous syndrome. Arch Cardiovasc Dis. (2020) 113:381–90. 10.1016/j.acvd.2020.03.012 [DOI] [PubMed] [Google Scholar]
- 44.Casebeer A, Horter L, Hayden J, Simmons J, Evers T. Phenotypic clustering of heart failure with preserved ejection fraction reveals different rates of hospitalization. J Cardiovasc Med. (2021) 22:45–52. 10.2459/jcm.0000000000001116 [DOI] [PubMed] [Google Scholar]
- 45.Woolley RJ, Ceelen D, Ouwerkerk W, Tromp J, Figarska SM, Anker SD, et al. Machine learning based on biomarker profiles identifies distinct subgroups of heart failure with preserved ejection fraction. Eur J Heart Fai. (2021) 23:983–91. 10.1002/ejhf.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedman ÅK, Hage C, Sharma A, Brosnan MJ, Buckbinder L, Gan LM, et al. Identification of novel pheno-groups in heart failure with preserved ejection fraction using machine learning. Heart. (2020) 106:342–9. 10.1136/heartjnl-2019-315481 [DOI] [PubMed] [Google Scholar]
- 47.Uijl A, Savarese G, Vaartjes I, Dahlström U, Brugts JJ, Linssen GCM, et al. Identification of distinct phenotypic clusters in heart failure with preserved ejection fraction. Eur J Heart Fail. (2021) 23:973–82. 10.1002/ejhf.2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn VS, Knutsdottir H, Luo X, Bedi K, Margulies KB, Haldar SM, et al. Myocardial gene expression signatures in human heart failure with preserved ejection fraction. Circulation. (2021) 143:120–34. 10.1161/circulationaha.120.050498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y, Wang H, Li Z, Cheng J, Fang R, Cao H, et al. Subtypes identification on heart failure with preserved ejection fraction via network enhancement fusion using multi-omics data. Comput Struct Biotechnol J. (2021) 19:1567–78. 10.1016/j.csbj.2021.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nouraei H, Rabkin SW. A new approach to the clinical subclassification of heart failure with preserved ejection fraction. Int J Cardiol. (2021) 331:138–43. 10.1016/j.ijcard.2021.01.052 [DOI] [PubMed] [Google Scholar]
- 51.Riolet C, Menet A, Verdun S, Altes A, Appert L, Guyomar Y, et al. Clinical and prognostic implications of phenomapping in patients with heart failure receiving cardiac resynchronization therapy. Arch Cardiovasc Dis. (2021) 114:197–210. 10.1016/j.acvd.2020.07.004 [DOI] [PubMed] [Google Scholar]
- 52.Bartko PE, Heitzinger G, Spinka G, Pavo N, Prausmüller S, Kastl S, et al. Principal morphomic and functional components of secondary mitral regurgitation. JACC Cardiovasc Imaging. (2021) 14:2288–300. 10.1016/j.jcmg.2021.05.020 [DOI] [PubMed] [Google Scholar]
- 53.Perry A, Loh F, Adamo L, Zhang KW, Deych E, Foraker R, et al. Unsupervised cluster analysis of patients with recovered left ventricular ejection fraction identifies unique clinical phenotypes. PLoS One. (2021) 16:e0248317. 10.1371/journal.pone.0248317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kao DP, Wagner BD, Robertson AD, Bristow MR, Lowes BD. A personalized BEST: characterization of latent clinical classes of nonischemic heart failure that predict outcomes and response to bucindolol. PLoS One. (2012) 7:e48184. 10.1371/journal.pone.0048184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith OR, Gidron Y, Kupper N, Winter JB, Denollet J. Vital exhaustion in chronic heart failure: symptom profiles and clinical outcome. J Psychosom Res. (2009) 66:195–201. 10.1016/j.jpsychores.2008.10.021 [DOI] [PubMed] [Google Scholar]
- 56.Lee KS, Song EK, Lennie TA, Frazier SK, Chung ML, Heo S, et al. Symptom clusters in men and women with heart failure and their impact on cardiac event-free survival. J Cardiovasc Nurs. (2010) 25:263–72. 10.1097/JCN.0b013e3181cfbb88 [DOI] [PubMed] [Google Scholar]
- 57.Sabbah MS, Fayyaz AU, de Denus S, Felker GM, Borlaug BA, Dasari S, et al. Obese-inflammatory phenotypes in heart failure with preserved ejection fraction. Circ Heart Fail. (2020) 13:e006414. 10.1161/circheartfailure.119.006414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segar MW, Patel KV, Ayers C, Basit M, Tang WHW, Willett D, et al. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur J Heart Fail. (2020) 22:148–58. 10.1002/ejhf.1621 [DOI] [PubMed] [Google Scholar]
- 59.Sethares KA, Viveiros JD, Ayotte B. Uncertainty levels differ by physical heart failure symptom cluster. Appl Nurs Res. (2021) 60:151435. [DOI] [PubMed] [Google Scholar]
- 60.Tromp J, Ouwerkerk W, Demissei BG, Anker SD, Cleland JG, Dickstein K, et al. Novel endotypes in heart failure: effects on guideline-directed medical therapy. Eur Heart J. (2018) 39:4269–76. 10.1093/eurheartj/ehy712 [DOI] [PubMed] [Google Scholar]
- 61.Zhao W-L, Deng C-H, Ngo C-W. k-means: a revisit. Neurocomputing. (2018) 291:195–206. 10.1016/j.neucom.2018.02.072 [DOI] [Google Scholar]
- 62.Towards Machine Learning. What is K-Means Algorithm and How It Works. Available online at: https://towardsmachinelearning.org/k-means/ [Google Scholar]
- 63.Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation. (2019) 139:e56–528. 10.1161/cir.0000000000000659 [DOI] [PubMed] [Google Scholar]
- 64.Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, et al. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. (2002) 347:1397–402. 10.1056/NEJMoa020265 [DOI] [PubMed] [Google Scholar]
- 65.Jackson SL, Tong X, King RJ, Loustalot F, Hong Y, Ritchey MD. National burden of heart failure events in the United States, 2006 to 2014. Circ Heart Fail. (2018) 11:e004873. 10.1161/circheartfailure.117.004873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma A, Zhao X, Hammill BG, Hernandez AF, Fonarow GC, Felker GM, et al. Trends in noncardiovascular comorbidities among patients hospitalized for heart failure: insights from the get with the guidelines-heart failure registry. Circ Heart Fail. (2018) 11:e004646. 10.1161/circheartfailure.117.004646 [DOI] [PubMed] [Google Scholar]
- 67.van Deursen VM, Urso R, Laroche C, Damman K, Dahlström U, Tavazzi L, et al. Co-morbidities in patients with heart failure: an analysis of the European Heart Failure Pilot Survey. Eu J Heart Fail. (2014) 16:103–11. 10.1002/ejhf.30 [DOI] [PubMed] [Google Scholar]
- 68.Mogensen UM, Ersbøll M, Andersen M, Andersson C, Hassager C, Torp-Pedersen C, et al. Clinical characteristics and major comorbidities in heart failure patients more than 85 years of age compared with younger age groups. Eur J Heart Fail. (2011) 13:1216–23. 10.1093/eurjhf/hfr116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.